1NSA
 
 | |
1NSB
 
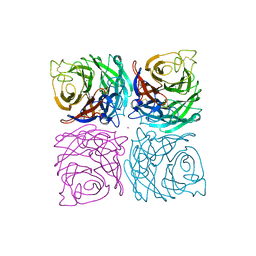 | |
1NSC
 
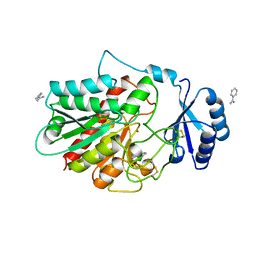
 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetyl-alpha-neuraminic acid, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
1NSD
 
 | | INFLUENZA B VIRUS NEURAMINIDASE CAN SYNTHESIZE ITS OWN INHIBITOR | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Burmeister, W.P, Ruigrok, R.W.H, Cusack, S. | | Deposit date: | 1993-05-24 | | Release date: | 1993-10-31 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Influenza B virus neuraminidase can synthesize its own inhibitor.
Structure, 1, 1993
|
|
1NSE
 
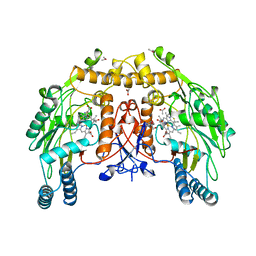 | | BOVINE ENDOTHELIAL NITRIC OXIDE SYNTHASE | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, CACODYLATE ION, ... | | Authors: | Raman, C.S, Li, H, Martasek, P, Kral, V, Masters, B.S.S, Poulos, T.L. | | Deposit date: | 1998-05-14 | | Release date: | 1999-05-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of constitutive endothelial nitric oxide synthase: a paradigm for pterin function involving a novel metal center.
Cell(Cambridge,Mass.), 95, 1998
|
|
1NSF
 
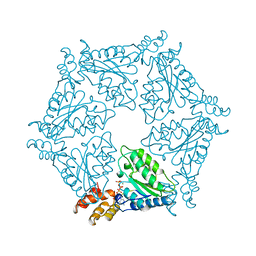 | | D2 HEXAMERIZATION DOMAIN OF N-ETHYLMALEIMIDE SENSITIVE FACTOR (NSF) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N-ETHYLMALEIMIDE SENSITIVE FACTOR | | Authors: | Yu, R.C, Hanson, P.I, Jahn, R, Brunger, A.T. | | Deposit date: | 1998-06-26 | | Release date: | 1998-11-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP.
Nat.Struct.Biol., 5, 1998
|
|
1NSG
 
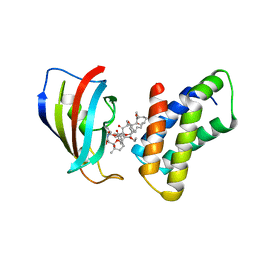 | | THE STRUCTURE OF THE IMMUNOPHILIN-IMMUNOSUPPRESSANT FKBP12-RAPAMYCIN COMPLEX INTERACTING WITH HUMAN FRAP | | Descriptor: | C49-METHYL RAPAMYCIN, FK506-BINDING PROTEIN, FKBP-RAPAMYCIN ASSOCIATED PROTEIN (FRAP) | | Authors: | Liang, J, Choi, J, Clardy, J. | | Deposit date: | 1997-07-01 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Refined structure of the FKBP12-rapamycin-FRB ternary complex at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1NSH
 
 | |
1NSI
 
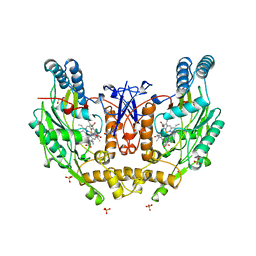 | | HUMAN INDUCIBLE NITRIC OXIDE SYNTHASE, ZN-BOUND, L-ARG COMPLEX | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ARGININE, GLYCEROL, ... | | Authors: | Li, H, Raman, C.S, Glaser, C.B, Blasko, E, Young, T.A, Parkinson, J.F, Whitlow, M, Poulos, T.L. | | Deposit date: | 1999-01-10 | | Release date: | 2000-01-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structures of zinc-free and -bound heme domain of human inducible nitric-oxide synthase. Implications for dimer stability and comparison with endothelial nitric-oxide synthase.
J.Biol.Chem., 274, 1999
|
|
1NSJ
 
 | |
1NSK
 
 | |
1NSL
 
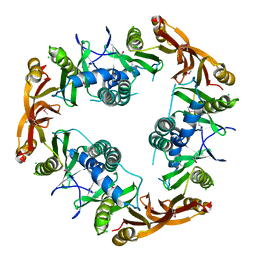 | | Crystal structure of Probable acetyltransferase | | Descriptor: | CHLORIDE ION, Probable acetyltransferase | | Authors: | Brunzelle, J.S, Korolev, S.V, Wu, R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-01-27 | | Release date: | 2003-07-29 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of Bacillus subtilis YdaF protein: A putative ribosomal N-acetyltransferase
Proteins, 57, 2004
|
|
1NSM
 
 | |
1NSN
 
 | |
1NSO
 
 | | Folded monomer of protease from Mason-Pfizer monkey virus | | Descriptor: | Protease 13 kDa | | Authors: | Veverka, V, Bauerova, H, Zabransky, A, Lang, J, Ruml, T, Pichova, I, Hrabal, R. | | Deposit date: | 2003-01-28 | | Release date: | 2003-02-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional structure of a monomeric form of a retroviral protease
J.MOL.BIOL., 333, 2003
|
|
1NSP
 
 | | MECHANISM OF PHOSPHATE TRANSFER BY NUCLEOSIDE DIPHOSPHATE KINASE: X-RAY STRUCTURES OF A PHOSPHO-HISTIDINE INTERMEDIATE OF THE ENZYMES FROM DROSOPHILA AND DICTYOSTELIUM | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Morera, S, Chiadmi, M, Lebras, G, Lascu, I. | | Deposit date: | 1995-04-18 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium.
Biochemistry, 34, 1995
|
|
1NSQ
 
 | | MECHANISM OF PHOSPHATE TRANSFER BY NUCLEOSIDE DIPHOSPHATE KINASE: X-RAY STRUCTURES OF A PHOSPHO-HISTIDINE INTERMEDIATE OF THE ENZYMES FROM DROSOPHILA AND DICTYOSTELIUM | | Descriptor: | NUCLEOSIDE DIPHOSPHATE KINASE | | Authors: | Janin, J, Chiadmi, M, Morera, S, Lebras, G, Lascu, I. | | Deposit date: | 1995-04-24 | | Release date: | 1995-07-10 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Mechanism of phosphate transfer by nucleoside diphosphate kinase: X-ray structures of the phosphohistidine intermediate of the enzymes from Drosophila and Dictyostelium.
Biochemistry, 34, 1995
|
|
1NSR
 
 | |
1NSS
 
 | |
1NST
 
 | |
1NSU
 
 | |
1NSV
 
 | |
1NSW
 
 | | The Crystal Structure of the K18G Mutant of the thioredoxin from Alicyclobacillus acidocaldarius | | Descriptor: | THIOREDOXIN | | Authors: | Bartolucci, S, De Simone, G, Galdiero, S, Improta, R, Menchise, V, Pedone, C, Pedone, E, Saviano, M. | | Deposit date: | 2003-01-28 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An integrated structural and computational study of the thermostability of two thioredoxin mutants from Alicyclobacillus acidocaldarius
J.Bacteriol., 185, 2003
|
|
1NSX
 
 | |
1NSY
 
 | | CRYSTAL STRUCTURE OF NH3-DEPENDENT NAD+ SYNTHETASE FROM BACILLUS SUBTILIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Rizzi, M, Nessi, C, Bolognesi, M, Galizzi, A, Coda, A. | | Deposit date: | 1996-07-22 | | Release date: | 1997-07-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NH3-dependent NAD+ synthetase from Bacillus subtilis.
EMBO J., 15, 1996
|
|
