1L8G
 
 | | Crystal structure of PTP1B complexed with 7-(1,1-Dioxo-1H-benzo[d]isothiazol-3-yloxymethyl)-2-(oxalyl-amino)-4,7-dihydro-5H-thieno[2,3-c]pyran-3-carboxylic acid | | Descriptor: | 7-(1,1-DIOXO-1H-BENZO[D]ISOTHIAZOL-3-YLOXYMETHYL)-2-(OXALYL-AMINO)-4,7-DIHYDRO-5H-THIENO[2,3-C]PYRAN-3-CARBOXYLIC ACID, PROTEIN-TYROSINE PHOSPHATASE, NON-RECEPTOR TYPE 1 | | Authors: | Iversen, L.F, Andersen, H.S, Moller, K.B, Olsen, O.H, Peters, G.H, Branner, S, Mortensen, S.B, Hansen, T.K, Lau, J, Ge, Y, Holsworth, D.D, Newman, M.J, Moller, N.P.H. | | Deposit date: | 2002-03-20 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Steric hindrance as a basis for structure-based design of selective inhibitors of protein-tyrosine phosphatases.
Biochemistry, 40, 2001
|
|
1L8H
 
 | | DNA PROTECTION AND BINDING BY E. COLI DPS PROTEIN | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, POTASSIUM ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2002-03-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1L8I
 
 | | Dna Protection and Binding by E. Coli DPS Protein | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA PROTECTION DURING STARVATION PROTEIN, POTASSIUM ION | | Authors: | Luo, J, Liu, D, White, M.A, Fox, R.O. | | Deposit date: | 2002-03-20 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | DNA Protection and Binding by E. Coli Dps Protein
To be Published
|
|
1L8J
 
 | | Crystal Structure of the Endothelial Protein C Receptor and Bound Phospholipid Molecule | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Endothelial protein C receptor, ... | | Authors: | Oganesyan, V, Oganesyan, N, Terzyan, S, Dongfeng, Q, Dauter, Z, Esmon, N.L, Esmon, C.T. | | Deposit date: | 2002-03-20 | | Release date: | 2002-06-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of the endothelial protein C receptor and a bound phospholipid.
J.Biol.Chem., 277, 2002
|
|
1L8K
 
 | | T Cell Protein-Tyrosine Phosphatase Structure | | Descriptor: | T-cell protein-tyrosine phosphatase | | Authors: | Iversen, L.F. | | Deposit date: | 2002-03-21 | | Release date: | 2002-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structure determination of T cell protein-tyrosine phosphatase.
J.Biol.Chem., 277, 2002
|
|
1L8L
 
 | | Molecular basis for the local confomational rearrangement of human phosphoserine phosphatase | | Descriptor: | D-2-AMINO-3-PHOSPHONO-PROPIONIC ACID, L-3-phosphoserine phosphatase | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase.
J.Biol.Chem., 277, 2002
|
|
1L8N
 
 | | The 1.5A crystal structure of alpha-D-glucuronidase from Bacillus stearothermophilus T-1, complexed with 4-O-methyl-glucuronic acid and xylotriose | | Descriptor: | 4-O-methyl-beta-D-glucopyranuronic acid, ALPHA-D-GLUCURONIDASE, GLYCEROL, ... | | Authors: | Golan, G, Shallom, D, Teplitsky, A, Zaide, G, Shulami, S, Baasov, T, Stojanoff, V, Thompson, A, Shoham, Y, Shoham, G. | | Deposit date: | 2002-03-21 | | Release date: | 2003-03-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structures of Geobacillus stearothermophilus {alpha}-Glucuronidase Complexed with Its Substrate and Products: MECHANISTIC IMPLICATIONS.
J.Biol.Chem., 279, 2004
|
|
1L8O
 
 | | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase | | Descriptor: | L-3-phosphoserine phosphatase, PHOSPHATE ION, SERINE | | Authors: | Kim, H.Y, Heo, Y.S, Kim, J.H, Park, M.H, Moon, J, Park, S.Y, Lee, T.G, Jeon, Y.H, Ro, S, Hwang, K.Y. | | Deposit date: | 2002-03-21 | | Release date: | 2003-04-01 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for the local conformational rearrangement of human phosphoserine phosphatase
J.Biol.Chem., 277, 2002
|
|
1L8P
 
 | | Mg-phosphonoacetohydroxamate complex of S39A yeast enolase 1 | | Descriptor: | MAGNESIUM ION, PHOSPHONOACETOHYDROXAMIC ACID, enolase 1 | | Authors: | Poyner, R.R, Larsen, T.M, Wong, S.W, Reed, G.H. | | Deposit date: | 2002-03-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional and structural changes due to a serine to alanine mutation in the active-site flap of enolase.
Arch.Biochem.Biophys., 401, 2002
|
|
1L8Q
 
 | |
1L8R
 
 | | Structure of the Retinal Determination Protein Dachshund Reveals a DNA-Binding Motif | | Descriptor: | Dachshund | | Authors: | Kim, S.S, Zhang, R, Braunstein, S.E, Joachimiak, A, Cvekl, A, Hegde, R.S. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of the retinal determination protein Dachshund reveals a DNA binding motif.
Structure, 10, 2002
|
|
1L8S
 
 | |
1L8T
 
 | |
1L8V
 
 | |
1L8W
 
 | | Crystal Structure of Lyme Disease Variable Surface Antigen VlsE of Borrelia burgdorferi | | Descriptor: | VlsE1 | | Authors: | Eicken, C, Sharma, V, Klabunde, T, Lawrenz, M.B, Hardham, J.M, Norris, S.J, Sacchettini, J.C. | | Deposit date: | 2002-03-21 | | Release date: | 2002-06-19 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Lyme disease variable surface antigen VlsE of Borrelia burgdorferi.
J.Biol.Chem., 277, 2002
|
|
1L8X
 
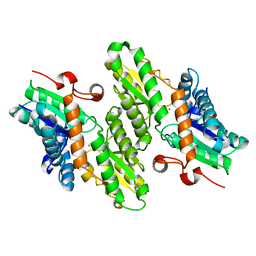 | | Crystal Structure of Ferrochelatase from the Yeast, Saccharomyces cerevisiae, with Cobalt(II) as the Substrate Ion | | Descriptor: | COBALT (II) ION, Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metal Binding to Saccharomyces cerevisiae Ferrochelatase
Biochemistry, 41, 2002
|
|
1L8Y
 
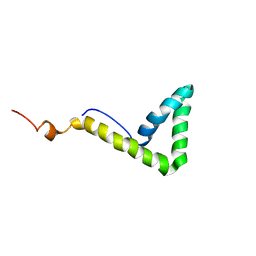 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
1L8Z
 
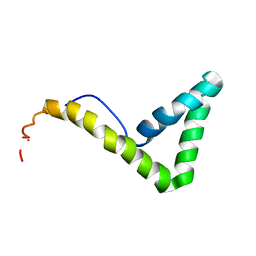 | | Solution structure of HMG box 5 in human upstream binding factor | | Descriptor: | upstream binding factor 1 | | Authors: | Yang, W, Xu, Y, Wu, J, Zeng, W, Shi, Y. | | Deposit date: | 2002-03-22 | | Release date: | 2002-06-05 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and DNA binding property of the fifth HMG box domain in comparison with the first HMG box domain in human upstream binding factor
Biochemistry, 42, 2003
|
|
1L90
 
 | |
1L91
 
 | |
1L92
 
 | |
1L93
 
 | |
1L94
 
 | |
1L95
 
 | |
1L96
 
 | | STRUCTURE OF A HINGE-BENDING BACTERIOPHAGE T4 LYSOZYME MUTANT, ILE3-> PRO | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Dixon, M, Shewchuk, L, Matthews, B.W. | | Deposit date: | 1992-02-11 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a hinge-bending bacteriophage T4 lysozyme mutant, Ile3-->Pro.
J.Mol.Biol., 227, 1992
|
|
