1KU2
 
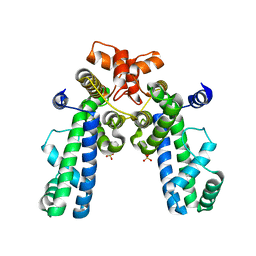 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment Containing Regions 1.2 to 3.1 | | Descriptor: | SULFATE ION, sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1KU3
 
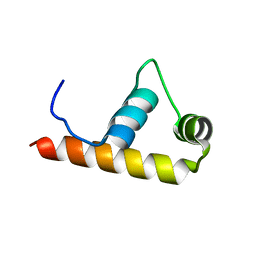 | | Crystal Structure of Thermus aquaticus RNA Polymerase Sigma Subunit Fragment, Region 4 | | Descriptor: | sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1KU5
 
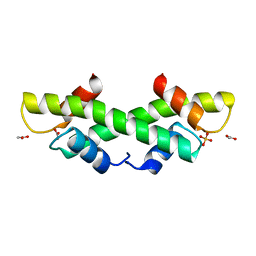 | | Crystal Structure of recombinant histone HPhA from hyperthermophilic archaeon Pyrococcus horikoshii OT3 | | Descriptor: | ACETATE ION, HPhA, SULFATE ION | | Authors: | Li, T, Sun, F, Ji, X, Feng, Y, Rao, Z. | | Deposit date: | 2002-01-21 | | Release date: | 2003-08-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure based hyperthermostability of archaeal histone HPhA from Pyrococcus horikoshii
J.MOL.BIOL., 325, 2003
|
|
1KU6
 
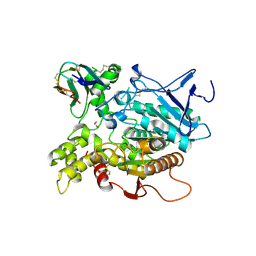 | | Fasciculin 2-Mouse Acetylcholinesterase Complex | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE, ... | | Authors: | Bourne, Y, Burmeister, W, Taylor, P, Marchot, P. | | Deposit date: | 2002-01-21 | | Release date: | 2003-12-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site.
EMBO J., 22, 2003
|
|
1KU7
 
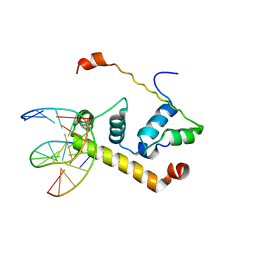 | | Crystal Structure of Thermus aquatics RNA Polymerase SigmaA Subunit Region 4 Bound to-35 Element DNA | | Descriptor: | 5'-D(*CP*CP*TP*TP*GP*AP*CP*AP*AP*AP*G)-3', 5'-D(*CP*CP*TP*TP*TP*GP*TP*CP*AP*AP*G)-3', sigma factor sigA | | Authors: | Campbell, E.A, Muzzin, O, Chlenov, M, Sun, J.L, Olson, C.A, Weinman, O, Trester-Zedlitz, M.L, Darst, S.A. | | Deposit date: | 2002-01-21 | | Release date: | 2002-03-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the bacterial RNA polymerase promoter specificity sigma subunit.
Mol.Cell, 9, 2002
|
|
1KU8
 
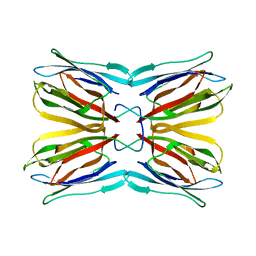 | | Crystal structure of Jacalin | | Descriptor: | JACALIN ALPHA CHAIN, JACALIN BETA CHAIN | | Authors: | Bourne, Y, Astoul, C.H, Zamboni, V, Peumans, W.J, Menu-Bouaouiche, L, Van Damme, E.J.M, Barre, A, Rouge, P. | | Deposit date: | 2002-01-21 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose.
Biochem.J., 364, 2002
|
|
1KU9
 
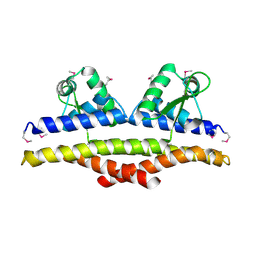 | | X-ray Structure of a Methanococcus jannaschii DNA-Binding Protein: Implications for Antibiotic Resistance in Staphylococcus aureus | | Descriptor: | hypothetical protein MJ223 | | Authors: | Ray, S.S, Bonanno, J.B, Chen, H, de Lencastre, H, Wu, S, Tomasz, A, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2002-01-21 | | Release date: | 2002-12-25 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | X-ray structure of an M. jannaschii DNA-binding protein: implications for antibiotic resistance in S.
aureus
Proteins, 50, 2002
|
|
1KUF
 
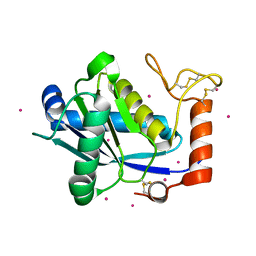 | | High-resolution Crystal Structure of a Snake Venom Metalloproteinase from Taiwan Habu | | Descriptor: | CADMIUM ION, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Yuann, J.M, Wang, A.H.J. | | Deposit date: | 2002-01-21 | | Release date: | 2002-07-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The 1.35 A structure of cadmium-substituted TM-3, a snake-venom metalloproteinase from Taiwan habu: elucidation of a TNFalpha-converting enzyme-like active-site structure with a distorted octahedral geometry of cadmium.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1KUG
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with its endogenous inhibitor pENW | | Descriptor: | CADMIUM ION, ENW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUH
 
 | | ZINC PROTEASE FROM STREPTOMYCES CAESPITOSUS | | Descriptor: | CALCIUM ION, ZINC ION, ZINC PROTEASE | | Authors: | Kurisu, G, Kinoshita, T, Sugimoto, A, Nagara, A, Kai, Y, Kasai, N, Harada, S. | | Deposit date: | 1996-02-22 | | Release date: | 1997-03-12 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of the zinc endoprotease from Streptomyces caespitosus.
J.Biochem.(Tokyo), 121, 1997
|
|
1KUI
 
 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEQW. | | Descriptor: | CADMIUM ION, EQW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUJ
 
 | | Crystal structure of Jacalin complexed with 1-O-methyl-alpha-D-mannose | | Descriptor: | JACALIN ALPHA CHAIN, JACALIN BETA CHAIN, methyl alpha-D-mannopyranoside | | Authors: | Bourne, Y, Astoul, C.H, Zamboni, V, Peumans, W.J, Menu-Bouaouiche, L, Van Damme, E.J.M, Barre, A, Rouge, P. | | Deposit date: | 2002-01-22 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the unusual carbohydrate-binding specificity of jacalin towards galactose and mannose.
Biochem.J., 364, 2002
|
|
1KUK
 
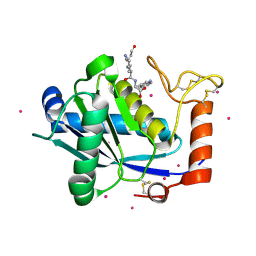 | | Crystal Structure of a Taiwan Habu Venom Metalloproteinase complexed with pEKW. | | Descriptor: | CADMIUM ION, EKW, metalloproteinase | | Authors: | Huang, K.F, Chiou, S.H, Ko, T.P, Wang, A.H.J. | | Deposit date: | 2002-01-22 | | Release date: | 2002-07-10 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Determinants of the inhibition of a Taiwan habu venom metalloproteinase by its endogenous inhibitors revealed by X-ray crystallography and synthetic inhibitor analogues.
Eur.J.Biochem., 269, 2002
|
|
1KUL
 
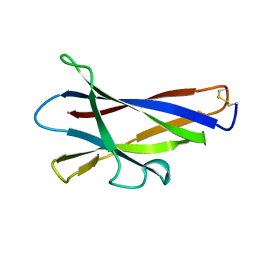 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, 5 STRUCTURES | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KUM
 
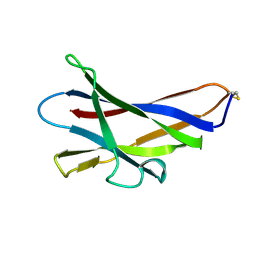 | | GLUCOAMYLASE, GRANULAR STARCH-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUCOAMYLASE | | Authors: | Sorimachi, K, Jacks, A.J, Le Gal-Coeffet, M.-F, Williamson, G, Archer, D.B, Williamson, M.P. | | Deposit date: | 1996-01-12 | | Release date: | 1996-07-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the granular starch binding domain of glucoamylase from Aspergillus niger by nuclear magnetic resonance spectroscopy.
J.Mol.Biol., 259, 1996
|
|
1KUN
 
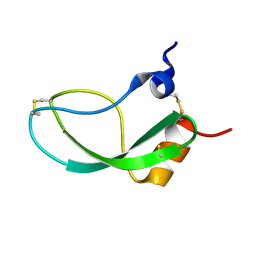 | | SOLUTION STRUCTURE OF THE HUMAN ALPHA3-CHAIN TYPE VI COLLAGEN C-TERMINAL KUNITZ DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | ALPHA3-CHAIN TYPE VI COLLAGEN | | Authors: | Sorensen, M.D, Bjorn, S, Norris, K, Olsen, O, Petersen, L, James, T.L, Led, J.J. | | Deposit date: | 1997-03-04 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure and backbone dynamics of the human alpha3-chain type VI collagen C-terminal Kunitz domain,.
Biochemistry, 36, 1997
|
|
1KUP
 
 | |
1KUQ
 
 | | CRYSTAL STRUCTURE OF T3C MUTANT S15 RIBOSOMAL PROTEIN IN COMPLEX WITH 16S RRNA | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, SULFATE ION | | Authors: | Nikulin, A.D, Tishchenko, S, Revtovich, S, Ehresmann, B, Ehresmann, C, Dumas, P, Garber, M, Nikonov, S, Nevskaya, N. | | Deposit date: | 2002-01-22 | | Release date: | 2003-06-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Role of N-terminal helix in interaction of ribosomal protein S15 with 16S rRNA.
Biochemistry Mosc., 69, 2004
|
|
1KUT
 
 | | Structural Genomics, Protein TM1243, (SAICAR synthetase) | | Descriptor: | Phosphoribosylaminoimidazole-succinocarboxamide synthase | | Authors: | Zhang, R, Skarina, T, Beasley, S, Edwards, A, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-22 | | Release date: | 2002-08-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of SAICAR synthase from Thermotoga maritima at 2.2 angstroms reveals an unusual covalent dimer.
Acta Crystallogr.,Sect.F, 62, 2006
|
|
1KUU
 
 | | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD | | Descriptor: | conserved protein | | Authors: | Saridakis, V, Christendat, D, Thygesen, A, Arrowsmith, C.H, Edwards, A.M, Pai, E.F. | | Deposit date: | 2002-01-22 | | Release date: | 2002-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | CRYSTAL STRUCTURE OF METHANOBACTERIUM THERMOAUTOTROPHICUM CONSERVED PROTEIN MTH1020 REVEALS AN NTN-HYDROLASE FOLD
PROTEINS: STRUCT.,FUNCT.,GENET., 48, 2002
|
|
1KUV
 
 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL 5-BROMOTRYPTAMINE, MAGNESIUM ION, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KUW
 
 | | High-Resolution Structure and Localization of Amylin Nucleation Site in Detergent Micelles | | Descriptor: | Islet amyloid polypeptide | | Authors: | Mascioni, A, Porcelli, F, Ilangovan, U, Ramamoorthy, A, Veglia, G. | | Deposit date: | 2002-01-22 | | Release date: | 2003-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Conformational preferences of the amylin nucleation site in SDS micelles: an NMR study.
Biopolymers, 69, 2003
|
|
1KUX
 
 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-TRIMETHYLENE-ACETYL-TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KUY
 
 | | X-ray Crystallographic Studies of Serotonin N-acetyltransferase Catalysis and Inhibition | | Descriptor: | COA-S-ACETYL TRYPTAMINE, Serotonin N-acetyltransferase | | Authors: | Wolf, E, De Angelis, J, Khalil, E.M, Cole, P.A, Burley, S.K. | | Deposit date: | 2002-01-22 | | Release date: | 2002-03-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray crystallographic studies of serotonin N-acetyltransferase catalysis and inhibition.
J.Mol.Biol., 317, 2002
|
|
1KUZ
 
 | |
