1KTB
 
 | | The Structure of alpha-N-Acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETIC ACID, ... | | Authors: | Garman, S.C, Hannick, L, Zhu, A, Garboczi, D.N. | | Deposit date: | 2002-01-15 | | Release date: | 2002-03-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The 1.9 A structure of alpha-N-acetylgalactosaminidase: molecular basis of glycosidase deficiency diseases
Structure, 10, 2002
|
|
1KTC
 
 | | The Structure of alpha-N-Acetylgalactosaminidase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Garman, S.C, Hannick, L, Zhu, A, Garboczi, D.N. | | Deposit date: | 2002-01-15 | | Release date: | 2002-03-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 1.9 A structure of alpha-N-acetylgalactosaminidase: molecular basis of glycosidase deficiency diseases.
Structure, 10, 2002
|
|
1KTD
 
 | | CRYSTAL STRUCTURE OF CLASS II MHC MOLECULE IEK BOUND TO PIGEON CYTOCHROME C PEPTIDE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion protein consisting of cytochrome C peptide, glycine rich linker, ... | | Authors: | Fremont, D.H, Dai, S, Chiang, H, Crawford, F, Marrack, P, Kappler, J. | | Deposit date: | 2002-01-15 | | Release date: | 2002-05-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of cytochrome c presentation by IE(k).
J.Exp.Med., 195, 2002
|
|
1KTE
 
 | |
1KTG
 
 | | Crystal Structure of a C. elegans Ap4A Hydrolase Binary Complex | | Descriptor: | ADENOSINE MONOPHOSPHATE, Diadenosine Tetraphosphate Hydrolase, HYDROXIDE ION, ... | | Authors: | Bailey, S, Sedelnikova, S.E, Blackburn, G.M, Abdelghany, H.M, Baker, P.J, McLennan, A.G, Rafferty, J.B. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of diadenosine tetraphosphate hydrolase from Caenorhabditis elegans in free and binary complex forms
Structure, 10, 2002
|
|
1KTH
 
 | |
1KTI
 
 | | BINDING OF 100 MM N-ACETYL-N'-BETA-D-GLUCOPYRANOSYL UREA TO GLYCOGEN PHOSPHORYLASE B: KINETIC AND CRYSTALLOGRAPHIC STUDIES | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-(acetylcarbamoyl)-beta-D-glucopyranosylamine, ... | | Authors: | Oikonomakos, N.G, Kosmopoulou, M, Zographos, S.E, Leonidas, D.D, Chrysina, E.D, Somsak, L, Nagy, V, Praly, J.P, Docsa, T, Toth, B, Gergely, P. | | Deposit date: | 2002-01-16 | | Release date: | 2002-01-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Binding of N-acetyl-N'-beta-D-glucopyranosyl urea and N-benzoyl-N'-beta-D-glucopyranosyl urea to glycogen phosphorylase b: kinetic and crystallographic studies.
Eur.J.Biochem., 269, 2002
|
|
1KTJ
 
 | | X-ray Structure Of Der P 2, The Major House Dust Mite Allergen | | Descriptor: | ALLERGEN DER P 2 | | Authors: | Derewenda, U, Li, J, Derewenda, Z, Dauter, Z, Mueller, G.A, Rule, G.S, Benjamin, D.C. | | Deposit date: | 2002-01-16 | | Release date: | 2002-05-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of a major dust mite allergen Der p 2, and its biological implications.
J.Mol.Biol., 318, 2002
|
|
1KTK
 
 | | Complex of Streptococcal pyrogenic enterotoxin C (SpeC) with a human T cell receptor beta chain (Vbeta2.1) | | Descriptor: | Exotoxin type C, T-cell receptor beta chain | | Authors: | Sundberg, E.J, Li, H, Llera, A.S, McCormick, J.K, Tormo, J, Karjalainen, K, Schlievert, P.M, Mariuzza, R.A. | | Deposit date: | 2002-01-16 | | Release date: | 2002-06-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of two streptococcal superantigens bound to TCR beta chains reveal diversity in the architecture of T cell signaling complexes.
Structure, 10, 2002
|
|
1KTL
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ALPHA CHAIN, ... | | Authors: | Holmes, M.A, Strong, R.K. | | Deposit date: | 2002-01-16 | | Release date: | 2003-02-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HLA-E allelic variants: Correlating differential expression, peptide affinities, crystal structures and thermal stabilities
J.Biol.Chem., 278, 2003
|
|
1KTM
 
 | |
1KTN
 
 | | Structural Genomics, Protein EC1535 | | Descriptor: | 2-deoxyribose-5-phosphate aldolase | | Authors: | Zhang, R, Joachimiak, A, Edwards, A, Skarina, T, Evdokimova, E, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-01-16 | | Release date: | 2002-08-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The 1.5A crystal structure of
2-deoxyribose-5-phosphate aldlase
To be Published
|
|
1KTO
 
 | |
1KTP
 
 | | Crystal structure of c-phycocyanin of synechococcus vulcanus at 1.6 angstroms | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA SUBUNIT, C-PHYCOCYANIN BETA SUBUNIT, ... | | Authors: | Adir, N, Dobrovetsky, E, Lerner, N. | | Deposit date: | 2002-01-17 | | Release date: | 2002-03-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Refined structure of c-phycocyanin from the cyanobacterium Synechococcus vulcanus at 1.6 A: insights into the role of solvent molecules in thermal stability and co-factor structure
Biochim.Biophys.Acta, 1556, 2002
|
|
1KTQ
 
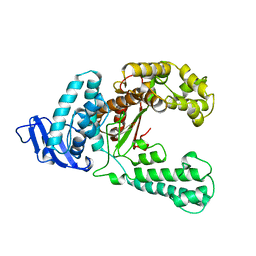 | | DNA POLYMERASE | | Descriptor: | DNA POLYMERASE I | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1995-08-16 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the large fragment of Thermus aquaticus DNA polymerase I at 2.5-A resolution: structural basis for thermostability.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
1KTR
 
 | | Crystal Structure of the Anti-His Tag Antibody 3D5 Single-Chain Fragment (scFv) in Complex with a Oligohistidine peptide | | Descriptor: | Anti-his tag antibody 3d5 variable light chain, Peptide linker, Anti-his tag antibody 3d5 variable heavy chain, ... | | Authors: | Kaufmann, M, Lindner, P, Honegger, A, Blank, K, Tschopp, M, Capitani, G, Plueckthun, A, Gruetter, M.G. | | Deposit date: | 2002-01-17 | | Release date: | 2002-05-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the anti-His tag antibody 3D5 single-chain fragment complexed to its antigen.
J.Mol.Biol., 318, 2002
|
|
1KTS
 
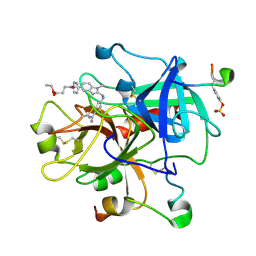 | | Thrombin Inhibitor Complex | | Descriptor: | 3-({2-[(4-CARBAMIMIDOYL-PHENYLAMINO)-METHYL]-3-METHYL-3H-BENZOIMIDAZOLE-5-CARBONYL}-PYRIDIN-2-YL-AMINO)-PROPIONIC ACID ETHYL ESTER, hirudin IIB, thrombin | | Authors: | Nar, H. | | Deposit date: | 2002-01-17 | | Release date: | 2002-02-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure-based design of novel potent nonpeptide thrombin inhibitors.
J.Med.Chem., 45, 2002
|
|
1KTT
 
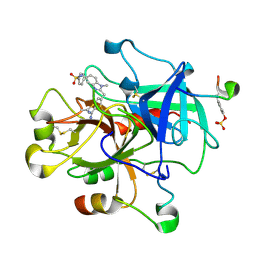 | | Thrombin inhibitor complex | | Descriptor: | 4-(5-BENZENESULFONYLAMINO-1-METHYL-1H-BENZOIMIDAZOL-2-YLMETHYL)-BENZAMIDINE, hirudin IIB, thrombin | | Authors: | Nar, H. | | Deposit date: | 2002-01-17 | | Release date: | 2002-02-06 | | Last modified: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based design of novel potent nonpeptide thrombin inhibitors.
J.Med.Chem., 45, 2002
|
|
1KTU
 
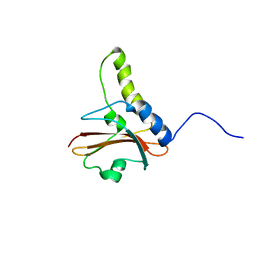 | | NuiA | | Descriptor: | NuiA | | Authors: | Kirby, T.W, Mueller, G.A, DeRose, E.F, Lebetkin, M.S, Meiss, G, Pingoud, A, London, R.E. | | Deposit date: | 2002-01-17 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The Nuclease A Inhibitor represents a new variation of the rare PR-1 fold.
J.Mol.Biol., 320, 2002
|
|
1KTV
 
 | |
1KTW
 
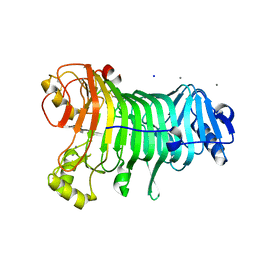 | | IOTA-CARRAGEENASE COMPLEXED TO IOTA-CARRAGEENAN FRAGMENTS | | Descriptor: | 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, 3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose-(1-4)-3,6-anhydro-2-O-sulfo-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Michel, G, Kahn, R, Dideberg, O. | | Deposit date: | 2002-01-18 | | Release date: | 2003-06-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structural Bases of the Processive Degradation of iota-Carrageenan, a Main Cell Wall Polysaccharide of Red Algae.
J.Mol.Biol., 334, 2003
|
|
1KTX
 
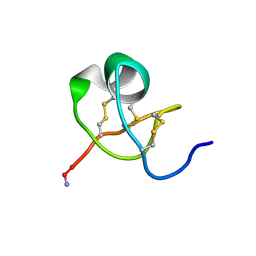 | | KALIOTOXIN (1-37) SHOWS STRUCTURAL DIFFERENCES WITH RELATED POTASSIUM CHANNEL BLOCKERS | | Descriptor: | KALIOTOXIN | | Authors: | Fernandez, I, Romi, R, Szendefi, S, Martin-Eauclaire, M.-F, Rochat, H, Van Rietschtoten, J, Pons, M, Giralt, E. | | Deposit date: | 1994-06-02 | | Release date: | 1995-01-26 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Kaliotoxin (1-37) shows structural differences with related potassium channel blockers.
Biochemistry, 33, 1994
|
|
1KTZ
 
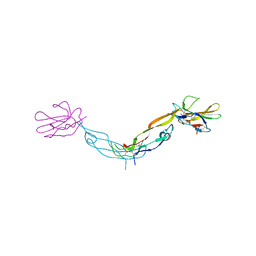 | | Crystal Structure of the Human TGF-beta Type II Receptor Extracellular Domain in Complex with TGF-beta3 | | Descriptor: | TGF-beta Type II Receptor, TRANSFORMING GROWTH FACTOR BETA 3 | | Authors: | Hart, P.J, Deep, S, Taylor, A.B, Shu, Z, Hinck, C.S, Hinck, A.P. | | Deposit date: | 2002-01-18 | | Release date: | 2002-02-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of the human TbetaR2 ectodomain--TGF-beta3 complex.
Nat.Struct.Biol., 9, 2002
|
|
1KU0
 
 | | Structure of the Bacillus stearothermophilus L1 lipase | | Descriptor: | CALCIUM ION, L1 lipase, ZINC ION | | Authors: | Jeong, S.-T, Kim, H.-K, Kim, S.-J, Chi, S.-W, Pan, J.-G, Oh, T.-K, Ryu, S.-E. | | Deposit date: | 2002-01-18 | | Release date: | 2002-08-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel zinc-binding center and a temperature switch in the Bacillus stearothermophilus L1 lipase.
J.Biol.Chem., 277, 2002
|
|
1KU1
 
 | | Crystal Structure of the Sec7 Domain of Yeast GEA2 | | Descriptor: | ARF guanine-nucleotide exchange factor 2 | | Authors: | Renault, L, Christova, P, Guibert, B, Pasqualato, S, Cherfils, J. | | Deposit date: | 2002-01-20 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Mechanism of domain closure of Sec7 domains and role in BFA sensitivity.
Biochemistry, 41, 2002
|
|
