1J3T
 
 | | Solution structure of the second SH3 domain of human intersectin 2 (KIAA1256) | | Descriptor: | Intersectin 2 | | Authors: | Nameki, N, Koshiba, S, Tochio, N, Kobayashi, N, Inoue, M, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-13 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second SH3 domain of human intersectin 2 (KIAA1256)
To be Published
|
|
1J3U
 
 | |
1J3W
 
 | |
1J3X
 
 | |
1J3Y
 
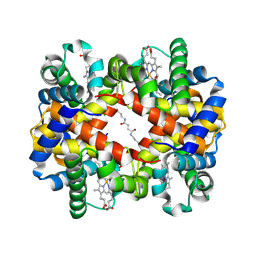 | | Direct observation of photolysis-induced tertiary structural changes in human hemoglobin; Crystal structure of alpha(Fe)-beta(Ni) hemoglobin (laser photolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1J3Z
 
 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Fe-CO)-beta(Ni) hemoglobin (laser unphotolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1J40
 
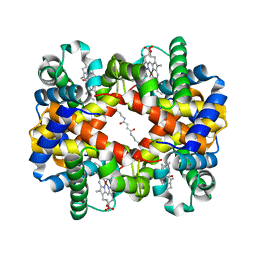 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Ni)-beta(Fe-CO) hemoglobin (laser unphotolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1J41
 
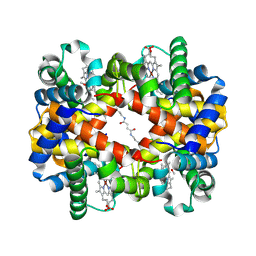 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Ni)-beta(Fe) hemoglobin (laser photolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1J42
 
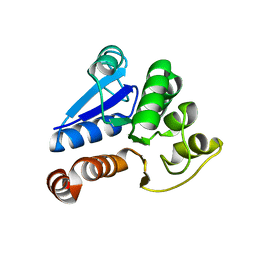 | | Crystal Structure of Human DJ-1 | | Descriptor: | RNA-binding protein regulatory subunit | | Authors: | Cha, S.S. | | Deposit date: | 2003-02-26 | | Release date: | 2004-02-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain.
J.Biol.Chem., 278, 2003
|
|
1J46
 
 | |
1J47
 
 | |
1J48
 
 | | Crystal Structure of Apo-C1027 | | Descriptor: | Apoprotein of C1027 | | Authors: | Chen, Y, Li, J, Liu, Y, Bartlam, M, Gao, Y, Jin, L, Tang, H, Shao, Y, Zhen, Y, Rao, Z. | | Deposit date: | 2001-07-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo-C1027 and Computer Modeling Analysis of C1027 Chromophore- Protein Complex
To be published
|
|
1J49
 
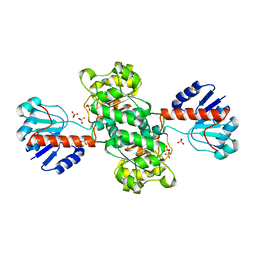
 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-14 | | Release date: | 2002-05-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
1J4A
 
 | | INSIGHTS INTO DOMAIN CLOSURE, SUBSTRATE SPECIFICITY AND CATALYSIS OF D-LACTATE DEHYDROGENASE FROM LACTOBACILLUS BULGARICUS | | Descriptor: | D-LACTATE DEHYDROGENASE, SULFATE ION | | Authors: | Razeto, A, Kochhar, S, Hottinger, H, Dauter, M, Wilson, K.S, Lamzin, V.S. | | Deposit date: | 2001-08-18 | | Release date: | 2002-05-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain closure, substrate specificity and catalysis of D-lactate dehydrogenase from Lactobacillus bulgaricus.
J.Mol.Biol., 318, 2002
|
|
1J4B
 
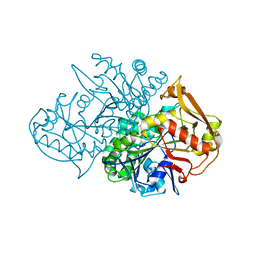 | | Recombinant Mouse-Muscle Adenylosuccinate Synthetase | | Descriptor: | adenylosuccinate synthetase | | Authors: | Iancu, C.V, Borza, T, Choe, J.Y, Fromm, H.J, Honzatko, R.B. | | Deposit date: | 2001-09-07 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Recombinant mouse muscle adenylosuccinate synthetase: overexpression, kinetics, and crystal structure.
J.Biol.Chem., 276, 2001
|
|
1J4E
 
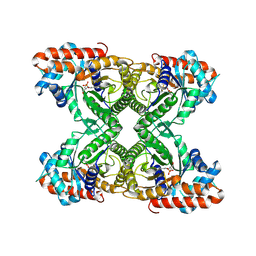 | | FRUCTOSE-1,6-BISPHOSPHATE ALDOLASE COVALENTLY BOUND TO THE SUBSTRATE DIHYDROXYACETONE PHOSPHATE | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, FRUCTOSE-BISPHOSPHATE ALDOLASE A | | Authors: | Choi, K.H, Shi, J, Hopkins, C.E, Tolan, D.R, Allen, K.N. | | Deposit date: | 2001-09-19 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Snapshots of catalysis: the structure of fructose-1,6-(bis)phosphate aldolase covalently bound to the substrate dihydroxyacetone phosphate.
Biochemistry, 40, 2001
|
|
1J4G
 
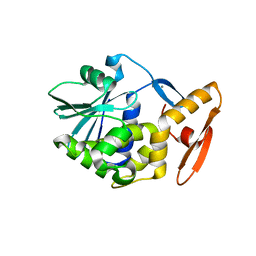 | | crystal structure analysis of the trichosanthin delta C7 | | Descriptor: | CALCIUM ION, Trichosanthin delta C7 | | Authors: | Ding, Y, Too, H.M, Wang, Z.L, Liu, Y.W, Dong, Y.C, Shaw, P.C, Rao, Z.H. | | Deposit date: | 2001-09-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | crystal structure analysis of the trichosanthin delta C7
To be Published
|
|
1J4H
 
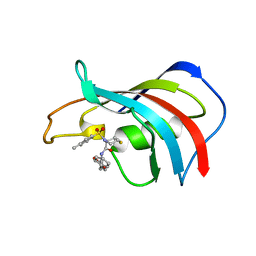 | | crystal structure analysis of the FKBP12 complexed with 000107 small molecule | | Descriptor: | 3-PHENYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PROPIONIC ACID ETHYL ESTER, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1J4I
 
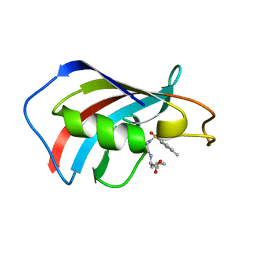 | | crystal structure analysis of the FKBP12 complexed with 000308 small molecule | | Descriptor: | 4-METHYL-2-{[4-(TOLUENE-4-SULFONYL)-THIOMORPHOLINE-3-CARBONYL]-AMINO}-PENTANOIC ACID, FKBP12 | | Authors: | Li, P, Ding, Y, Wang, L, Wu, B, Shu, C, Li, S, Shen, B, Rao, Z. | | Deposit date: | 2001-09-30 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design and structure-based study of new potential FKBP12 inhibitors.
Biophys.J., 85, 2003
|
|
1J4J
 
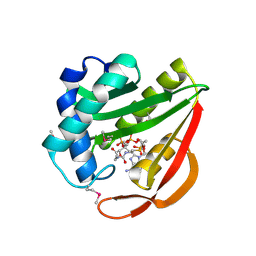 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
1J4K
 
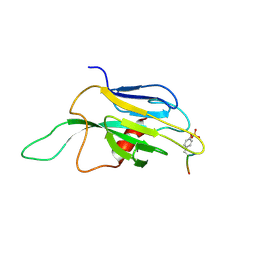 | |
1J4L
 
 | |
1J4M
 
 | | Minimized average structure of the 14-residue peptide RG-KWTY-NG-ITYE-GR (MBH12) | | Descriptor: | MBH12 | | Authors: | Pastor, M.T, Lopez de la Paz, M, Lacroix, E, Serrano, L, Perez-Paya, E. | | Deposit date: | 2001-10-10 | | Release date: | 2001-10-17 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Combinatorial approaches: a new tool to search for highly structured beta-hairpin peptides.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1J4N
 
 | | Crystal Structure of the AQP1 water channel | | Descriptor: | AQUAPORIN 1, nonyl beta-D-glucopyranoside | | Authors: | Sui, H, Han, B.-G, Lee, J.K, Walian, P, Jap, B.K. | | Deposit date: | 2001-10-19 | | Release date: | 2002-03-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of water-specific transport through the AQP1 water channel.
Nature, 414, 2001
|
|
1J4O
 
 | | REFINED NMR STRUCTURE OF THE FHA1 DOMAIN OF YEAST RAD53 | | Descriptor: | PROTEIN KINASE SPK1 | | Authors: | Yuan, C, Yongkiettrakul, S, Byeon, I.-J.L, Zhou, S, Tsai, M.-D. | | Deposit date: | 2001-10-03 | | Release date: | 2001-12-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structures of two FHA1-phosphothreonine peptide complexes provide insight into the structural basis of the ligand specificity of FHA1 from yeast Rad53.
J.Mol.Biol., 314, 2001
|
|
