1I3N
 
 | | MOLECULAR BASIS FOR SEVERE EPIMERASE-DEFICIENCY GALACTOSEMIA: X-RAY STRUCTURE OF THE HUMAN V94M-SUBSTITUTED UDP-GALACTOSE 4-EPIMERASE | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Thoden, J.B, Wohlers, T.M, Fridovich-Keil, J.L, Holden, H.M. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4-epimerase.
J.Biol.Chem., 276, 2001
|
|
1I3O
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF XIAP-BIR2 AND CASPASE 3 | | Descriptor: | BACULOVIRAL IAP REPEAT-CONTAINING PROTEIN 4, CASPASE 3, ZINC ION | | Authors: | Riedl, S.J, Renatus, M, Schwarzenbacher, R, Zhou, Q, Sun, C, Fesik, S.W, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2001-02-15 | | Release date: | 2001-03-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for the inhibition of caspase-3 by XIAP.
Cell(Cambridge,Mass.), 104, 2001
|
|
1I3P
 
 | |
1I3Q
 
 | | RNA POLYMERASE II CRYSTAL FORM I AT 3.1 A RESOLUTION | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II 13.6KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.2KD POLYPEPTIDE, DNA-DIRECTED RNA POLYMERASE II 14.5KD POLYPEPTIDE, ... | | Authors: | Cramer, P, Bushnell, D.A, Kornberg, R.D. | | Deposit date: | 2001-02-15 | | Release date: | 2001-04-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution.
Science, 292, 2001
|
|
1I3R
 
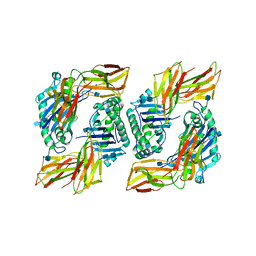 | | CRYSTAL STRUCTURE OF A MUTANT IEK CLASS II MHC MOLECULE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, FUSION PROTEIN CONSISTING OF MHC E-BETA-K PRECURSOR, ... | | Authors: | Kappler, J.W, Wilson, N. | | Deposit date: | 2001-02-15 | | Release date: | 2001-06-20 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Mutations changing the kinetics of class II MHC peptide exchange.
Immunity, 14, 2001
|
|
1I3S
 
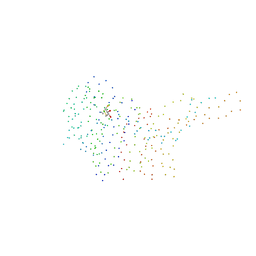 | | THE 2.7 ANGSTROM RESOLUTION CRYSTAL STRUCTURE OF A MUTATED BACULOVIRUS P35 AFTER CASPASE CLEAVAGE | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, EARLY 35 KDA PROTEIN | | Authors: | dela Cruz, W.P, Lemongello, D, Friesen, P.D, Fisher, A.J. | | Deposit date: | 2001-02-15 | | Release date: | 2001-10-24 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of baculovirus P35 reveals a novel conformational change in the reactive site loop after caspase cleavage.
J.Biol.Chem., 276, 2001
|
|
1I3T
 
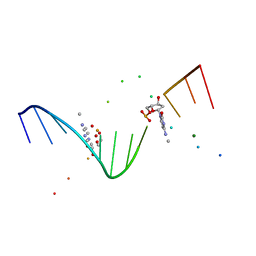 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATT(MO4)CGCG): THE WATSON-CRICK TYPE AND WOBBLE N4-METHOXYCYTIDINE/GUANOSINE BASE PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(C45)P*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Hossain, M.T, Hikima, T, Chatake, T, Masaru, T, Sunami, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs: III. N(4)-methoxycytosine can form both Watson-Crick type and wobbled base pairs in a B-form duplex.
J.Biochem.(Tokyo), 130, 2001
|
|
1I3U
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LLAMA VHH DOMAIN COMPLEXED WITH THE DYE RR1 | | Descriptor: | 5-(4,6-DIAMINO-[1,3,5]TRIAZIN-2-YLAMINO)-4-HYDROXY-3-(2-SULFO-PHENYLAZO)-NAPHTHALENE-2,7-DISULFONIC ACID, ANTIBODY VHH LAMA DOMAIN, SULFATE ION | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
1I3V
 
 | | THREE-DIMENSIONAL STRUCTURE OF A LAMA VHH DOMAIN UNLIGANDED | | Descriptor: | ANTIBODY VHH LAMA DOMAIN | | Authors: | Spinelli, S, Tegoni, M, Frenken, L, van Vliet, C, Cambillau, C. | | Deposit date: | 2001-02-16 | | Release date: | 2001-08-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Lateral recognition of a dye hapten by a llama VHH domain.
J.Mol.Biol., 311, 2001
|
|
1I3W
 
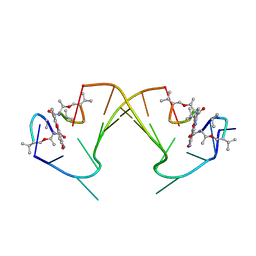 | | ACTINOMYCIN D BINDING TO CGATCGATCG | | Descriptor: | 5'-D(*C*GP*AP*TP*CP*GP*AP*(BRU)P*CP*GP)-3', ACTINOMYCIN D | | Authors: | Robinson, H, Gao, Y.-G, Yang, X.-L, Sanishvili, R, Joachimiak, A, Wang, A.H.-J. | | Deposit date: | 2001-02-17 | | Release date: | 2001-05-21 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystallographic Analysis of a Novel Complex of Actinomycin D Bound to the DNA Decamer Cgatcgatcg.
Biochemistry, 40, 2001
|
|
1I3X
 
 | |
1I3Y
 
 | |
1I3Z
 
 | | MURINE EAT2 SH2 DOMAIN IN COMPLEX WITH SLAM PHOSPHOPEPTIDE | | Descriptor: | EWS/FLI1 ACTIVATED TRANSCRIPT 2, SIGNALING LYMPHOCYTIC ACTIVATION MOLECULE | | Authors: | Lu, J, Poy, F, Morra, M, Terhorst, C, Eck, M.J. | | Deposit date: | 2001-02-19 | | Release date: | 2003-04-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the interaction of the free SH2 domain EAT-2 with
SLAM receptors in hematopoietic cells.
Eur.J.Biochem., 20, 2001
|
|
1I40
 
 | | STRUCTURE OF INORGANIC PYROPHOSPHATASE | | Descriptor: | CALCIUM ION, CHLORIDE ION, INORGANIC PYROPHOSPHATASE, ... | | Authors: | Samygina, V.R, Popov, A.N, Lamzin, V.S, Avaeva, S.M. | | Deposit date: | 2001-02-19 | | Release date: | 2001-12-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The structures of Escherichia coli inorganic pyrophosphatase complexed with Ca(2+) or CaPP(i) at atomic resolution and their mechanistic implications.
J.Mol.Biol., 314, 2001
|
|
1I41
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR APPA | | Descriptor: | 2-[(3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YLMETHYL)-IMINO]-5-PHOSPHONO-PENT-3-ENOIC ACID, CYSTATHIONINE GAMMA-SYNTHASE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1I42
 
 | | NMR STRUCTURE OF THE UBX DOMAIN FROM P47 | | Descriptor: | P47 | | Authors: | Yuan, X, Shaw, A, Zhang, X, Kondo, H, Lally, J, Freemont, P.S, Matthews, S. | | Deposit date: | 2001-02-19 | | Release date: | 2001-08-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and interaction surface of the C-terminal domain from p47: a major p97-cofactor involved in SNARE disassembly.
J.Mol.Biol., 311, 2001
|
|
1I43
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR PPCA | | Descriptor: | 3-(PHOSPHONOMETHYL)PYRIDINE-2-CARBOXYLIC ACID, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-19 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1I44
 
 | | CRYSTALLOGRAPHIC STUDIES OF AN ACTIVATION LOOP MUTANT OF THE INSULIN RECEPTOR TYROSINE KINASE | | Descriptor: | INSULIN RECEPTOR, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Till, J.H, Ablooglu, A.J, Frankel, M, Kohanski, R.A, Hubbard, S.R. | | Deposit date: | 2001-02-19 | | Release date: | 2001-03-07 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystallographic and solution studies of an activation loop mutant of the insulin receptor tyrosine kinase: insights into kinase mechanism.
J.Biol.Chem., 276, 2001
|
|
1I45
 
 | | YEAST TRIOSEPHOSPHATE ISOMERASE (MUTANT) | | Descriptor: | TRIOSEPHOSPHATE ISOMERASE | | Authors: | Rozovsky, S, Jogl, G, Tong, L, McDermott, A.E. | | Deposit date: | 2001-02-19 | | Release date: | 2001-06-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Solution-state NMR investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics.
J.Mol.Biol., 310, 2001
|
|
1I46
 
 | |
1I47
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATT(MO4)CGCG): THE WATSON-CRICK TYPE AND WOBBLE N4-METHOXYCYTIDINE/GUANOSINE BASE PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(C45)P*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Hossain, M.T, Hikima, T, Chatake, T, Tsunoda, M, Sunami, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-02-20 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on damaged DNAs: III. N(4)-methoxycytosine can form both Watson-Crick type and wobbled base pairs in a B-form duplex
J.Biochem.(Tokyo), 130, 2001
|
|
1I48
 
 | | CYSTATHIONINE GAMMA-SYNTHASE IN COMPLEX WITH THE INHIBITOR CTCPO | | Descriptor: | CARBOXYMETHYLTHIO-3-(3-CHLOROPHENYL)-1,2,4-OXADIAZOL, CYSTATHIONINE GAMMA-SYNTHASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Steegborn, C, Laber, B, Messerschmidt, A, Huber, R, Clausen, T. | | Deposit date: | 2001-02-20 | | Release date: | 2001-09-05 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structures of cystathionine gamma-synthase inhibitor complexes rationalize the increased affinity of a novel inhibitor.
J.Mol.Biol., 311, 2001
|
|
1I49
 
 | | CRYSTAL STRUCTURE ANALYSIS OF ARFAPTIN | | Descriptor: | ARFAPTIN 2 | | Authors: | Tarricone, C, Xiao, B, Justin, N, Gamblin, S.J, Smerdon, S.J. | | Deposit date: | 2001-02-20 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways.
Nature, 411, 2001
|
|
1I4A
 
 | | CRYSTAL STRUCTURE OF PHOSPHORYLATION-MIMICKING MUTANT T6D OF ANNEXIN IV | | Descriptor: | ANNEXIN IV, CALCIUM ION, SULFATE ION | | Authors: | Kaetzel, M.A, Mo, Y.D, Mealy, T.R, Campos, B, Bergsma-Schutter, W, Brisson, A, Dedman, J.R, Seaton, B.A. | | Deposit date: | 2001-02-20 | | Release date: | 2001-04-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphorylation mutants elucidate the mechanism of annexin IV-mediated membrane aggregation.
Biochemistry, 40, 2001
|
|
1I4B
 
 | |
