1CT6
 
 | | SOLUTION STRUCTURE OF CGGIRGERG IN CONTACT WITH THE MONOCLONAL ANTIBODY MAB 4X11, NMR, 11 STRUCTURES | | Descriptor: | HISTONE H3 PEPTIDE | | Authors: | Phan Chan Du, A, Petit, M.C, Guichard, G, Briand, J.P, Muller, S, Cung, M.T. | | Deposit date: | 1999-08-19 | | Release date: | 1999-09-02 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of antibody-bound peptides and retro-inverso analogues. A transferred nuclear Overhauser effect spectroscopy and molecular dynamics approach.
Biochemistry, 40, 2001
|
|
1CT8
 
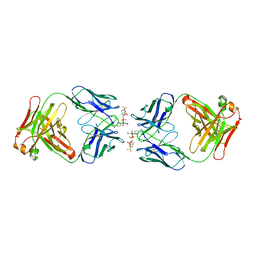 | | CATALYTIC ANTIBODY 7C8 COMPLEX | | Descriptor: | 7C8 FAB FRAGMENT; LONG CHAIN, 7C8 FAB FRAGMENT; SHORT CHAIN, SULFATE ION, ... | | Authors: | Gigant, B, Tsumuraya, T, Fujii, I, Knossow, M. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Diverse structural solutions to catalysis in a family of antibodies.
Structure Fold.Des., 7, 1999
|
|
1CT9
 
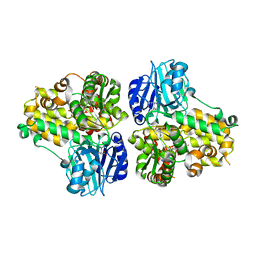 | | CRYSTAL STRUCTURE OF ASPARAGINE SYNTHETASE B FROM ESCHERICHIA COLI | | Descriptor: | ADENOSINE MONOPHOSPHATE, ASPARAGINE SYNTHETASE B, CHLORIDE ION, ... | | Authors: | Larsen, T.M, Boehlein, S.K, Schuster, S.M, Richards, N.G.J, Thoden, J.B, Holden, H.M, Rayment, I. | | Deposit date: | 1999-08-20 | | Release date: | 1999-12-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three-dimensional structure of Escherichia coli asparagine synthetase B: a short journey from substrate to product.
Biochemistry, 38, 1999
|
|
1CTA
 
 | |
1CTD
 
 | |
1CTE
 
 | |
1CTF
 
 | |
1CTI
 
 | |
1CTJ
 
 | | CRYSTAL STRUCTURE OF CYTOCHROME C6 | | Descriptor: | CYTOCHROME C6, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sheldrick, G.M. | | Deposit date: | 1995-08-08 | | Release date: | 1996-06-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ab initio determination of the crystal structure of cytochrome c6 and comparison with plastocyanin.
Structure, 3, 1995
|
|
1CTL
 
 | | STRUCTURE OF THE CARBOXY-TERMINAL LIM DOMAIN FROM THE CYSTEINE RICH PROTEIN CRP | | Descriptor: | AVIAN CYSTEINE RICH PROTEIN, ZINC ION | | Authors: | Perez-Alvarado, G.C, Miles, C, Michelsen, J.W, Louis, H.A, Winge, D.R, Beckerle, M.C, Summers, M.F. | | Deposit date: | 1995-01-06 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the carboxy-terminal LIM domain from the cysteine rich protein CRP.
Nat.Struct.Biol., 1, 1994
|
|
1CTM
 
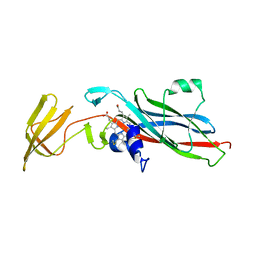 | | CRYSTAL STRUCTURE OF CHLOROPLAST CYTOCHROME F REVEALS A NOVEL CYTOCHROME FOLD AND UNEXPECTED HEME LIGATION | | Descriptor: | CYTOCHROME F, HEME C | | Authors: | Martinez, S.E, Huang, D, Szczepaniak, A, Cramer, W.A, Smith, J.L. | | Deposit date: | 1994-01-02 | | Release date: | 1994-05-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of chloroplast cytochrome f reveals a novel cytochrome fold and unexpected heme ligation.
Structure, 2, 1994
|
|
1CTN
 
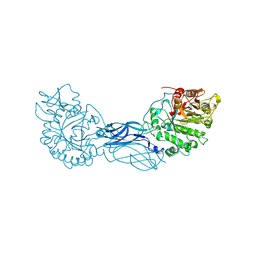 | | CRYSTAL STRUCTURE OF A BACTERIAL CHITINASE AT 2.3 ANGSTROMS RESOLUTION | | Descriptor: | CHITINASE A | | Authors: | Perrakis, A, Tews, I, Dauter, Z, Wilson, K.S, Vorgias, C.E. | | Deposit date: | 1994-10-10 | | Release date: | 1995-02-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bacterial chitinase at 2.3 A resolution.
Structure, 2, 1994
|
|
1CTO
 
 | | NMR STRUCTURE OF THE C-TERMINAL DOMAIN OF THE LIGAND-BINDING REGION OF MURINE GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRANULOCYTE COLONY-STIMULATING FACTOR RECEPTOR | | Authors: | Yamasaki, K, Naito, S, Anaguchi, H, Ohkubo, T, Ota, Y. | | Deposit date: | 1996-09-25 | | Release date: | 1997-10-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of an extracellular domain containing the WSxWS motif of the granulocyte colony-stimulating factor receptor and its interaction with ligand.
Nat.Struct.Biol., 4, 1997
|
|
1CTP
 
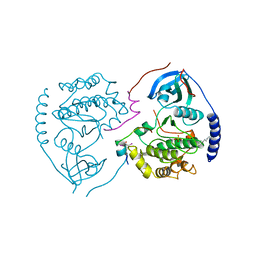 | | STRUCTURE OF THE MAMMALIAN CATALYTIC SUBUNIT OF CAMP-DEPENDENT PROTEIN KINASE AND AN INHIBITOR PEPTIDE DISPLAYS AN OPEN CONFORMATION | | Descriptor: | MYRISTIC ACID, cAMP-DEPENDENT PROTEIN KINASE, cAMP-dependent protein kinase inhibitor, ... | | Authors: | Karlsson, R, Zheng, J, Xuong, N.H, Taylor, S.S, Sowadski, J.M. | | Deposit date: | 1993-04-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the mammalian catalytic subunit of cAMP-dependent protein kinase and an inhibitor peptide displays an open conformation.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1CTQ
 
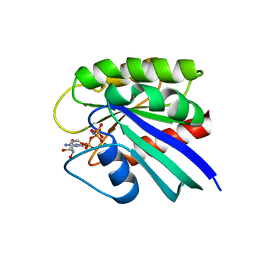 | | STRUCTURE OF P21RAS IN COMPLEX WITH GPPNHP AT 100 K | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, PROTEIN (TRANSFORMING PROTEIN P21/H-RAS-1) | | Authors: | Scheidig, A, Burmester, C, Goody, R.S. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | The pre-hydrolysis state of p21(ras) in complex with GTP: new insights into the role of water molecules in the GTP hydrolysis reaction of ras-like proteins.
Structure Fold.Des., 7, 1999
|
|
1CTR
 
 | |
1CTS
 
 | |
1CTT
 
 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 3,4-DIHYDRO-1H-PYRIMIDIN-2-ONE NUCLEOSIDE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
1CTU
 
 | | TRANSITION-STATE SELECTIVITY FOR A SINGLE OH GROUP DURING CATALYSIS BY CYTIDINE DEAMINASE | | Descriptor: | 4-HYDROXY-3,4-DIHYDRO-ZEBULARINE, CYTIDINE DEAMINASE, ZINC ION | | Authors: | Xiang, S, Short, S.A, Wolfenden, R, Carter, C.W. | | Deposit date: | 1995-02-11 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Transition-state selectivity for a single hydroxyl group during catalysis by cytidine deaminase.
Biochemistry, 34, 1995
|
|
1CTW
 
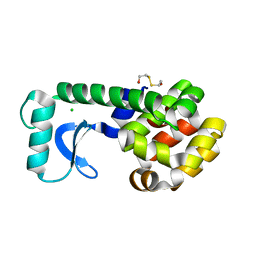 | | T4 LYSOZYME MUTANT I78A | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CTX
 
 | |
1CTY
 
 | |
1CTZ
 
 | |
1CU0
 
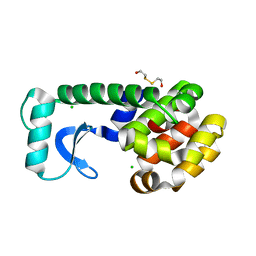 | | T4 LYSOZYME MUTANT I78M | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, LYSOZYME | | Authors: | Gassner, N.C, Baase, W.A, Lindstrom, J.D, Lu, J, Matthews, B.W. | | Deposit date: | 1999-08-20 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Methionine and alanine substitutions show that the formation of wild-type-like structure in the carboxy-terminal domain of T4 lysozyme is a rate-limiting step in folding.
Biochemistry, 38, 1999
|
|
1CU1
 
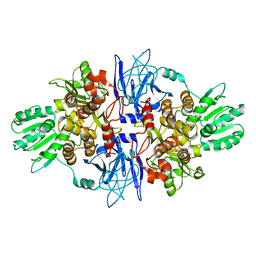 | | CRYSTAL STRUCTURE OF AN ENZYME COMPLEX FROM HEPATITIS C VIRUS | | Descriptor: | PHOSPHATE ION, PROTEIN (PROTEASE/HELICASE NS3), ZINC ION | | Authors: | Yao, N, Weber, P.C. | | Deposit date: | 1999-08-20 | | Release date: | 2000-08-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase.
Structure Fold.Des., 7, 1999
|
|
