1DLA
 
 | | NOVEL NADPH-BINDING DOMAIN REVEALED BY THE CRYSTAL STRUCTURE OF ALDOSE REDUCTASE | | Descriptor: | ALDOSE REDUCTASE | | Authors: | Rondeau, J.-M, Tete-Favier, F, Podjarny, A, Reymann, J.-M, Barth, P, Biellmann, J.-F, Moras, D. | | Deposit date: | 1993-02-08 | | Release date: | 1994-04-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Novel NADPH-binding domain revealed by the crystal structure of aldose reductase.
Nature, 355, 1992
|
|
8I41
 
 | |
8IO5
 
 | | Herg1a-herg1b closed state 2 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
8IO3
 
 | | Cryo-EM structure of human HCN3 channel with cilobradine | | Descriptor: | 3-[[(3~{S})-1-[2-(3,4-dimethoxyphenyl)ethyl]piperidin-3-yl]methyl]-7,8-dimethoxy-2,5-dihydro-1~{H}-3-benzazepin-4-one, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM structure of human HCN3 channel with cilobradine
To Be Published
|
|
1DKA
 
 | | DIALKYLGLYCINE DECARBOXYLASE STRUCTURE: BIFUNCTIONAL ACTIVE SITE AND ALKALI METAL BINDING SITES | | Descriptor: | 2,2-DIALKYLGLYCINE DECARBOXYLASE (PYRUVATE), 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, POTASSIUM ION, ... | | Authors: | Toney, M.D, Hohenester, E, Jansonius, J.N. | | Deposit date: | 1993-06-18 | | Release date: | 1994-10-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dialkylglycine decarboxylase structure: bifunctional active site and alkali metal sites.
Science, 261, 1993
|
|
7WGC
 
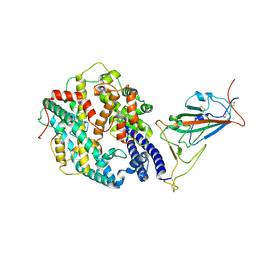 | | Neutral Omicron Spike Trimer in complex with ACE2. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Last modified: | 2022-07-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
7WG7
 
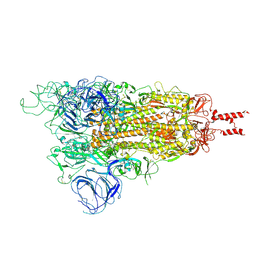 | | Acidic Omicron Spike Trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
8INZ
 
 | | Cryo-EM structure of human HCN3 channel in apo state | | Descriptor: | 4-[[(2~{S},4~{a}~{R},6~{S},8~{a}~{S})-6-[(4~{S},5~{R})-4-[(2~{S})-butan-2-yl]-5,9-dimethyl-decyl]-4~{a}-methyl-2,3,4,5,6,7,8,8~{a}-octahydro-1~{H}-naphthalen-2-yl]oxy]-4-oxidanylidene-butanoic acid, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 3 | | Authors: | Yu, B, Lu, Q.Y, Li, J, Zhang, J. | | Deposit date: | 2023-03-10 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | Cryo-EM structure of human HCN3 channel and its regulation by cAMP.
J.Biol.Chem., 300, 2024
|
|
1DP4
 
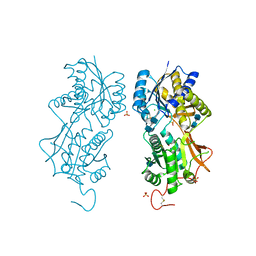 | | DIMERIZED HORMONE BINDING DOMAIN OF THE ATRIAL NATRIURETIC PEPTIDE RECEPTOR | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ATRIAL NATRIURETIC PEPTIDE RECEPTOR A, ... | | Authors: | van den Akker, F, Zhang, X, Miyagi, M, Huo, X, Misono, K.S, Yee, V.C. | | Deposit date: | 1999-12-23 | | Release date: | 2000-07-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the dimerized hormone-binding domain of a guanylyl-cyclase-coupled receptor.
Nature, 406, 2000
|
|
8IN7
 
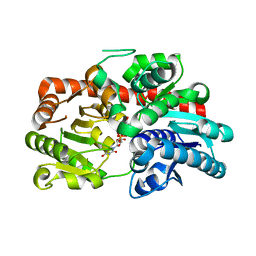 | | Crystal structure of UGT74AN3-UDP | | Descriptor: | Glycosyltransferase, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Feng, L, Wei, H. | | Deposit date: | 2023-03-08 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of UGT74AN3-UDP
To Be Published
|
|
8IOB
 
 | | Herg1a-herg1b closed state 1 | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Herg1a-herg1b closed state 1
To Be Published
|
|
8I0X
 
 | | Beta-Xylosidase JB13GH39P28 showing salt/ethanol/trypsin tolerance, low-pH/low-Temperature activity, and transformation of notoginsenosides | | Descriptor: | Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | beta-Xylosidase JB13GH39P28(D41G)showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
1ESC
 
 | | THE MOLECULAR MECHANISM OF ENANTIORECOGNITION BY ESTERASES | | Descriptor: | ESTERASE | | Authors: | Wei, Y, Schottel, J.L, Derewenda, U, Swenson, L, Patkar, S, Derewenda, Z.S. | | Deposit date: | 1994-10-07 | | Release date: | 1995-10-15 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A novel variant of the catalytic triad in the Streptomyces scabies esterase.
Nat.Struct.Biol., 2, 1995
|
|
8I42
 
 | |
1EFD
 
 | | PERIPLASMIC FERRIC SIDEROPHORE BINDING PROTEIN FHUD COMPLEXED WITH GALLICHROME | | Descriptor: | FERRICHROME-BINDING PERIPLASMIC PROTEIN, GALLICHROME | | Authors: | Clarke, T.E, Ku, S.-Y, Dougan, D.R, Vogel, H.J, Tari, L.W. | | Deposit date: | 2000-02-07 | | Release date: | 2000-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structure of the ferric siderophore binding protein FhuD complexed with gallichrome.
Nat.Struct.Biol., 7, 2000
|
|
7WGB
 
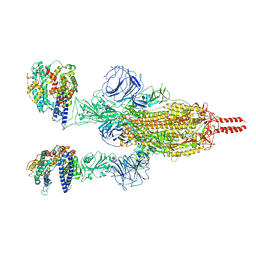 | | Neutral Omicron Spike Trimer in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Cui, Z. | | Deposit date: | 2021-12-28 | | Release date: | 2022-06-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural and functional characterizations of infectivity and immune evasion of SARS-CoV-2 Omicron.
Cell, 185, 2022
|
|
8I48
 
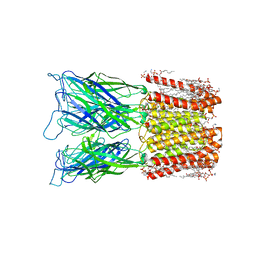 | | Cryo-EM structure of nanodisc (PE:PS:PC) reconstituted GLIC at pH 4 in closed state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHLORIDE ION, Proton-gated ion channel | | Authors: | Bharambe, N, Li, Z, Basak, S. | | Deposit date: | 2023-01-18 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cryo-EM structures of prokaryotic ligand-gated ion channel GLIC provide insights into gating in a lipid environment.
Nat Commun, 15, 2024
|
|
8I47
 
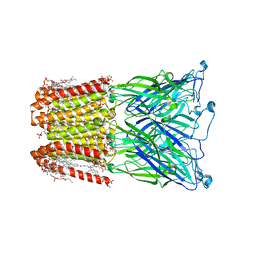 | |
1EIE
 
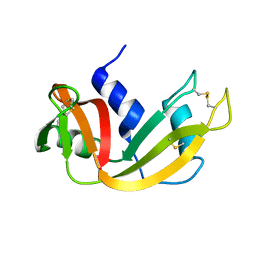 | |
8IOG
 
 | |
8IO4
 
 | | Herg1a-herg1b open state | | Descriptor: | Potassium voltage-gated channel subfamily H member 2 | | Authors: | Zhang, M.F. | | Deposit date: | 2023-03-10 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of Herg1a-herg1b open state
To Be Published
|
|
8I0J
 
 | | JB13GH39P28 mutant-D41G | | Descriptor: | CHLORIDE ION, Glycoside hydrolase family 39 beta-xylosidase | | Authors: | Zhou, J.P, Cao, L.J, Lin, M.Y, Zhang, R, Huang, Z.X. | | Deposit date: | 2023-01-11 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | beta-Xylosidase JB13GH39P28 (D41G) showing salt/ethanol/trypsin tolerance and transformation of notoginsenosides
To Be Published
|
|
7WLW
 
 | | X-ray structure of thermostabilized Drosophila dopamine transporter with GABA transporter1-like substitutions in the binding site, in complex with SKF89976a | | Descriptor: | (3S)-1-(4,4-diphenylbut-3-enyl)piperidine-3-carboxylic acid, Antibody Fragment heavy chain, Antibody fragment light chain, ... | | Authors: | Penmatsa, A, Joseph, D. | | Deposit date: | 2022-01-13 | | Release date: | 2022-06-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into GABA transport inhibition using an engineered neurotransmitter transporter.
Embo J., 41, 2022
|
|
1EJ4
 
 | | COCRYSTAL STRUCTURE OF EIF4E/4E-BP1 PEPTIDE | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE, EUKARYOTIC INITIATION FACTOR 4E, EUKARYOTIC TRANSLATION INITIATION FACTOR 4E BINDING PROTEIN 1 | | Authors: | Marcotrigiano, J, Gingras, A.-C, Sonenberg, N, Burley, S.K. | | Deposit date: | 2000-02-28 | | Release date: | 2000-03-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cap-dependent translation initiation in eukaryotes is regulated by a molecular mimic of eIF4G.
Mol.Cell, 3, 1999
|
|
1EET
 
 | | HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH THE INHIBITOR MSC204 | | Descriptor: | 1-(5-BROMO-PYRIDIN-2-YL)-3-[2-(6-FLUORO-2-HYDROXY-3-PROPIONYL-PHENYL)-CYCLOPROPYL]-UREA, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Hogberg, M, Sahlberg, C, Engelhardt, P, Noreen, R, Kangasmetsa, J, Johansson, N.G, Oberg, B, Vrang, L, Zhang, H, Sahlberg, B.L, Unge, T, Lovgren, S, Fridborg, K, Backbro, K. | | Deposit date: | 2000-02-03 | | Release date: | 2001-02-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Urea-PETT compounds as a new class of HIV-1 reverse transcriptase inhibitors. 3. Synthesis and further structure-activity relationship studies of PETT analogues.
J.Med.Chem., 42, 1999
|
|
