7Z9P
 
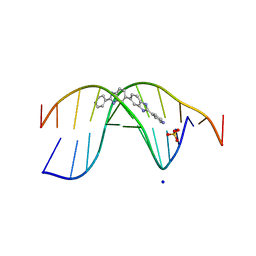 | | The novel DNA binding mechanism of ridinilazole, a precision Clostridiodes difficile antibiotic | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G*(RID))-3'), Ridinilazole, SODIUM ION, ... | | Authors: | Mason, S, Leonard, P.M. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Novel DNA Binding Mechanism of Ridinilazole, a Precision Clostridiodes difficile Antibiotic.
Antimicrob.Agents Chemother., 67, 2023
|
|
8RGC
 
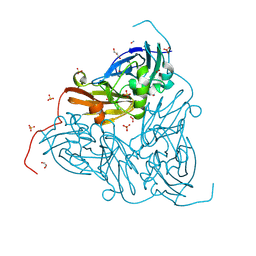 | | High pH (8.0) nitrite-bound MSOX movie series dataset 10 of the copper nitrite reductase from Bradyrhizobium sp. ORS375 (two-domain) [6.8 MGy] | | Descriptor: | CARBON DIOXIDE, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Rose, S.L, Ferroni, F.M, Antonyuk, S.V, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
6BIX
 
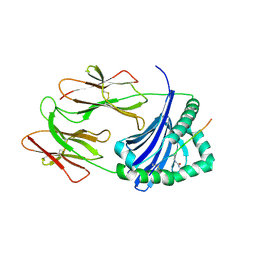 | | HLA-DRB1 in complex with citrullinated LL37 peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HLA class II DR-beta (HLA-DR B), HLA class II histocompatibility antigen, ... | | Authors: | Ting, Y.T, Scally, S.W, Rossjohn, J. | | Deposit date: | 2017-11-03 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The interplay between citrullination and HLA-DRB1 polymorphism in shaping peptide binding hierarchies in rheumatoid arthritis.
J. Biol. Chem., 293, 2018
|
|
7AAJ
 
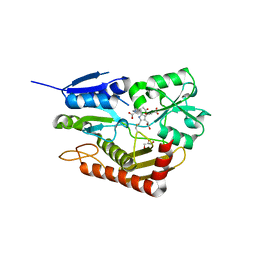 | | Human porphobilinogen deaminase in complex with cofactor | | Descriptor: | 3-[5-{[3-(2-carboxyethyl)-4-(carboxymethyl)-5-methyl-1H-pyrrol-2-yl]methyl}-4-(carboxymethyl)-1H-pyrrol-3-yl]propanoic acid, GLYCEROL, Porphobilinogen deaminase | | Authors: | Kallio, J.P, Bustad, H.J, Martinez, A. | | Deposit date: | 2020-09-04 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Characterization of porphobilinogen deaminase mutants reveals that arginine-173 is crucial for polypyrrole elongation mechanism.
Iscience, 24, 2021
|
|
7A7Z
 
 | |
8RSU
 
 | |
7Z81
 
 | | REP-related Chom18 variant with double TC mismatch | | Descriptor: | Chom18-TC DNA, STRONTIUM ION | | Authors: | Svoboda, J, Schneider, B, Berdar, D, Kolenko, P. | | Deposit date: | 2022-03-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformation-based refinement of 18-mer DNA structures.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
7LKU
 
 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3b (deuterated analog of inhibitor 2a) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7A8L
 
 | |
7PXO
 
 | | Structure of the Diels Alderase enzyme AbyU, from Micromonospora maris, co-crystallised with a non transformable substrate analogue | | Descriptor: | (2~{S},4~{S})-1-(4-methoxy-5-methyl-2-oxidanylidene-3~{H}-furan-3-yl)-2,4-dimethyl-dodecane-1,5-dione, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, YD repeat-containing protein | | Authors: | Back, C.R, Race, P.R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Delineation of the Complete Reaction Cycle of a Natural Diels-Alderase
To Be Published
|
|
7LKS
 
 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2f | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,2S,4S)-bicyclo[2.2.1]heptan-2-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7AA1
 
 | | Structural comparison of cellular retinoic acid binding proteins I and II in the presence and absence of natural and synthetic ligands | | Descriptor: | 4-[2-(5,5,8,8-tetramethyl-6,7-dihydroquinoxalin-2-yl)ethynyl]benzoic acid, Cellular retinoic acid-binding protein 2 | | Authors: | Tomlinson, C.W.E, Cornish, K.A.S, Pohl, E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structure-functional relationship of cellular retinoic acid-binding proteins I and II interacting with natural and synthetic ligands.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7LKV
 
 | | 1.55 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3c | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3R,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 1,2-ETHANEDIOL, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
6ZN4
 
 | | MaeB malic enzyme domain apoprotein | | Descriptor: | MAGNESIUM ION, NADP-dependent malate dehydrogenase,Malate dehydrogenase | | Authors: | Lovering, A.L, Harding, C.J. | | Deposit date: | 2020-07-06 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A rotary mechanism for allostery in bacterial hybrid malic enzymes.
Nat Commun, 12, 2021
|
|
6I5X
 
 | | Crystal structure of Aspergillus fumigatus phosphomannomutase | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, ... | | Authors: | Zhang, Y, Raimi, O.G, Ferenbach, A.T, van Aalten, D.M.F. | | Deposit date: | 2018-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Aspergillus fumigatus phosphomannomutase
To Be Published
|
|
7A7S
 
 | | rsGreenF in the green-on state | | Descriptor: | Green fluorescent protein | | Authors: | De Zitter, E, Dedecker, P, Van Meervelt, L. | | Deposit date: | 2020-08-30 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Function Dataset Reveals Environment Effects within a Fluorescent Protein Model System*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7LKT
 
 | | 1.50 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2k | | Descriptor: | (1R,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((adamantan-1-ylmethoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7LKR
 
 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 2a | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1R,2S)-2-((S)-2-(((((1R,3s,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3r,5S)-bicyclo[3.3.1]nonan-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
7A8N
 
 | |
8RZK
 
 | | The Michaelis complex of ZgGH129 D486N from Zobellia galactanivorans with neo-b/k-oligo-carrageenan tetrasaccharide (beta-kappa neo-oligo-carrageenan DP4). | | Descriptor: | 1,2-ETHANEDIOL, 3,6-anhydro-alpha-D-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-D-galactopyranose-(1-3)-4-O-sulfo-beta-D-galactopyranose, CHLORIDE ION, ... | | Authors: | Roret, T, Czjzek, M, Ficko-Blean, E. | | Deposit date: | 2024-02-12 | | Release date: | 2024-07-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Constrained Catalytic Itinerary of a Retaining 3,6-Anhydro-D-Galactosidase, a Key Enzyme in Red Algal Cell Wall Degradation.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6BND
 
 | | Crystal structure of the intrinsic colistin resistance enzyme ICR(Mc) from Moraxella catarrhalis, catalytic domain, Thr315Ala mutant mono-zinc and phosphoethanolamine complex | | Descriptor: | PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER, POLYETHYLENE GLYCOL (N=34), Phosphoethanolamine transferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Wawrzak, Z, Savchenko, A, Anderson, W.F, Satchell, K.J, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-16 | | Release date: | 2018-01-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Substrate Recognition by a Colistin Resistance Enzyme from Moraxella catarrhalis.
ACS Chem. Biol., 13, 2018
|
|
7LKW
 
 | | 1.65 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 3d (deuterated analog of inhibitor 3c) | | Descriptor: | (1R,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, (1S,2S)-2-((S)-2-(((((1R,3S,5S)-bicyclo[3.3.1]non-6-en-3-yl)methoxy)carbonyl)amino)-4-methylpentanamido)-1-hydroxy-3-((S)-2-oxopyrrolidin-3-yl)propane-1-sulfonic acid, 3C-like proteinase, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Chamandi, S.D, Rathnayake, A.D, Nguyen, H.N, Baird, M.A, Kim, Y, Shadipeni, N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2021-02-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Guided Design of Conformationally Constrained Cyclohexane Inhibitors of Severe Acute Respiratory Syndrome Coronavirus-2 3CL Protease.
J.Med.Chem., 64, 2021
|
|
6I1I
 
 | | Crystal structure of TP domain from Escherichia coli penicillin-binding protein 3 in complex with penicillin | | Descriptor: | Peptidoglycan D,D-transpeptidase FtsI,Peptidoglycan D,D-transpeptidase FtsI, Piperacillin (Open Form) | | Authors: | Bellini, D, Koekemoer, L, Newman, H, Dowson, C.G. | | Deposit date: | 2018-10-28 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Novel and Improved Crystal Structures of H. influenzae, E. coli and P. aeruginosa Penicillin-Binding Protein 3 (PBP3) and N. gonorrhoeae PBP2: Toward a Better Understanding of beta-Lactam Target-Mediated Resistance.
J.Mol.Biol., 431, 2019
|
|
7A85
 
 | |
8S5X
 
 | | High pH (8.0) as-isolated MSOX movie series dataset 10 of the copper nitrite reductase (NirK) from Bradyrhizobium japonicum USDA110 [3.5 MGy] | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, SULFATE ION, ... | | Authors: | Rose, S.L, Ferroni, F.M, Horrell, S, Brondino, C.D, Eady, R.R, Jaho, S, Hough, M.A, Owen, A.L, Antonyuk, S.V, Hasnain, S.S. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Spectroscopically Validated pH-dependent MSOX Movies Provide Detailed Mechanism of Copper Nitrite Reductases.
J.Mol.Biol., 436, 2024
|
|
