1GD3
 
 | | refined solution structure of human cystatin A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix
conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
1HG9
 
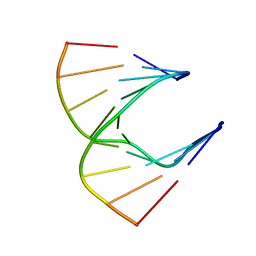 | | Solution structure of DNA:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*TP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-13 | | Release date: | 2002-01-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity.
Chemistry, 6, 2000
|
|
2NC9
 
 | | Apo solution structure of Hop TPR2A | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Darby, J.F, Vidler, L.R, Simpson, P.J, Matthews, S.J, Sharp, S.Y, Pearl, L.H, Hoelder, S, Workman, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hop TPR2A domain and investigation of target druggability by NMR, biochemical and in silico approaches.
Sci Rep, 10, 2020
|
|
1HIC
 
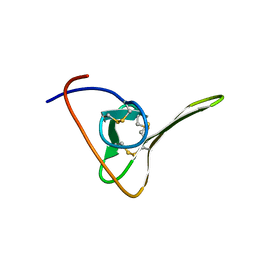 | |
1HHW
 
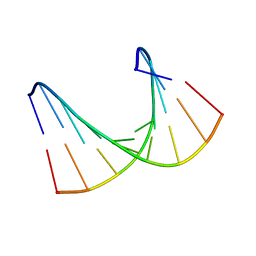 | | Solution structure of LNA1:RNA hybrid | | Descriptor: | 5- D(*CP*TP*GP*AP*+TLNP*AP*TP*GP*C) -3, 5- R(*GP*CP*AP*UP*AP*UP*CP*AP*G) -3 | | Authors: | Petersen, M, Bondensgaard, K, Wengel, J, Jacobsen, J.P. | | Deposit date: | 2000-12-29 | | Release date: | 2002-05-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Locked Nucleic Acid (Lna) Recognition of RNA: NMR Solution Structures of Lna:RNA Hybrids
J.Am.Chem.Soc., 124, 2002
|
|
1JIC
 
 | |
1T0K
 
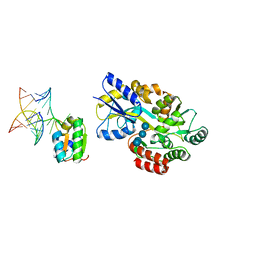 | | Joint X-ray and NMR Refinement of Yeast L30e-mRNA complex | | Descriptor: | 5'-R(*G*GP*AP*CP*GP*CP*AP*GP*AP*GP*AP*UP*GP*GP*UP*C)-3', 5'-R(*GP*AP*CP*CP*GP*GP*AP*GP*UP*GP*UP*CP*C)-3', 60S ribosomal protein L30, ... | | Authors: | Chao, J.A, Williamson, J.R. | | Deposit date: | 2004-04-09 | | Release date: | 2004-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Joint X-Ray and NMR Refinement of the Yeast L30e-mRNA Complex
Structure, 12, 2004
|
|
1VRV
 
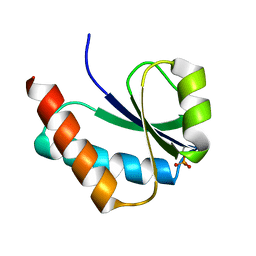 | |
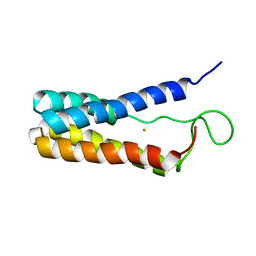
1EKY
 
 | |
2RRL
 
 | | Solution structure of the C-terminal domain of the FliK | | Descriptor: | Flagellar hook-length control protein | | Authors: | Mizuno, S, Tate, S, Kobayashi, N, Amida, H. | | Deposit date: | 2011-01-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The NMR structure of FliK, the trigger for the switch of substrate specificity in the flagellar type III secretion apparatus
J.Mol.Biol., 409, 2011
|
|
1FJC
 
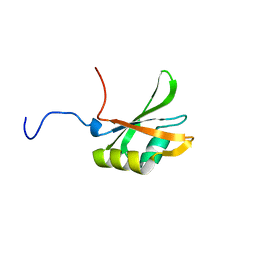 | |
1CNP
 
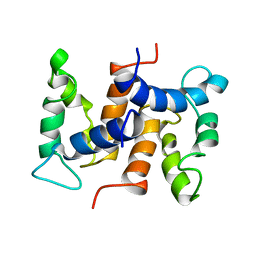 | | THE STRUCTURE OF CALCYCLIN REVEALS A NOVEL HOMODIMERIC FOLD FOR S100 CA2+-BINDING PROTEINS, NMR, 22 STRUCTURES | | Descriptor: | CALCYCLIN (RABBIT, APO) | | Authors: | Potts, B.C.M, Smith, J, Akke, M, Macke, T.J, Okazaki, K, Hidaka, H, Case, D.A, Chazin, W.J. | | Deposit date: | 1995-08-31 | | Release date: | 1996-10-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of calcyclin reveals a novel homodimeric fold for S100 Ca(2+)-binding proteins.
Nat.Struct.Biol., 2, 1995
|
|
1FJ7
 
 | |
1GIW
 
 | | SOLUTION STRUCTURE OF REDUCED HORSE HEART CYTOCHROME C, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Banci, L, Bertini, I, Huber, J.G, Spyroulias, G.A, Turano, P. | | Deposit date: | 1998-06-17 | | Release date: | 1998-12-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of reduced horse heart cytochrome c.
J.Biol.Inorg.Chem., 4, 1999
|
|
2WQQ
 
 | | Crystallographic analysis of monomeric CstII | | Descriptor: | ALPHA-2,3-/2,8-SIALYLTRANSFERASE, CYTIDINE-5'-MONOPHOSPHATE-3-FLUORO-N-ACETYL-NEURAMINIC ACID, DI(HYDROXYETHYL)ETHER | | Authors: | Chan, P.H.W, Lairson, L.L, Lee, H.J, Wakarchuk, W.W, Strynadka, N.C.J, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2009-08-25 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | NMR Spectroscopic Characterization of the Sialyltransferase Cstii from Camplyobacter Jejuni: Histidine 188 is the General Base.
Biochemistry, 48, 2009
|
|
1GD4
 
 | | SOLUTION STRUCTURE OF P25S CYSTATIN A | | Descriptor: | CYSTATIN A | | Authors: | Shimba, N, Kariya, E, Tate, S, Kaji, H, Kainosho, M. | | Deposit date: | 2000-09-08 | | Release date: | 2001-09-08 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structural comparison between wild-type and P25S human cystatin A by NMR spectroscopy. Does this mutation affect the a-helix conformation ?
J.STRUCT.FUNCT.GENOM., 1, 2000
|
|
2WCC
 
 | | phage lambda IntDBD1-64 complex with p prime 2 DNA | | Descriptor: | DNA (5'-D(*DCP*DGP*DAP*DGP*DTP*DCP*DAP *DAP*DAP*DAP*DTP*DC)-3'), DNA (5'-D(*DGP*DAP*DTP*DTP*DTP*DTP*DGP *DAP*DCP*DTP*DGP*DC)-3'), INTEGRASE | | Authors: | Fadeev, E.A, Sam, M.D, Clubb, R.T. | | Deposit date: | 2009-03-11 | | Release date: | 2009-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Amino-Terminal Domain of the Lambda Integrase Protein in Complex with DNA: Immobilization of a Flexible Tail Facilitates Beta- Sheet Recognition of the Major Groove.
J.Mol.Biol., 388, 2009
|
|
1DVH
 
 | | STRUCTURE AND DYNAMICS OF FERROCYTOCHROME C553 FROM DESULFOVIBRIO VULGARIS STUDIED BY NMR SPECTROSCOPY AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | CYTOCHROME C553, HEME C | | Authors: | Blackledge, M.J, Medvedeva, S, Poncin, M, Guerlesquin, F, Bruschi, M, Marion, D. | | Deposit date: | 1995-02-24 | | Release date: | 1995-06-03 | | Last modified: | 2021-03-03 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of ferrocytochrome c553 from Desulfovibrio vulgaris studied by NMR spectroscopy and restrained molecular dynamics.
J.Mol.Biol., 245, 1995
|
|
2OPV
 
 | |
1GBQ
 
 | | SOLUTION NMR STRUCTURE OF THE GRB2 N-TERMINAL SH3 DOMAIN COMPLEXED WITH A TEN-RESIDUE PEPTIDE DERIVED FROM SOS DIRECT REFINEMENT AGAINST NOES, J-COUPLINGS, AND 1H AND 13C CHEMICAL SHIFTS, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GRB2, SOS-1 | | Authors: | Wittekind, M, Mapelli, C, Lee, V, Goldfarb, V, Friedrichs, M.S, Meyers, C.A, Mueller, L. | | Deposit date: | 1996-12-23 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts.
J.Mol.Biol., 267, 1997
|
|
1HDN
 
 | |
1ZWT
 
 | | Structure of the globular head domain of the bundlin, BfpA, of the bundle-forming pilus of Enteropathogenic E.coli | | Descriptor: | Major structural subunit of bundle-forming pilus | | Authors: | Ramboarina, S, Fernandes, P.J, Daniell, S, Islam, S, Frankel, G, Booy, F, Donnenberg, M.S, Matthews, S. | | Deposit date: | 2005-06-06 | | Release date: | 2005-10-04 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Structure of the Bundle-forming Pilus from Enteropathogenic Escherichia coli
J.Biol.Chem., 280, 2005
|
|
2MTE
 
 | | Solution structure of Doc48S | | Descriptor: | CALCIUM ION, Cellulose 1,4-beta-cellobiosidase (reducing end) CelS | | Authors: | Chen, C, Feng, Y. | | Deposit date: | 2014-08-18 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Revisiting the NMR solution structure of the Cel48S type-I dockerin module from Clostridium thermocellum reveals a cohesin-primed conformation.
J.Struct.Biol., 188, 2014
|
|
1JTJ
 
 | | Solution structure of HIV-1Lai mutated SL1 hairpin | | Descriptor: | HIV-1Lai SL1 | | Authors: | Kieken, F, Arnoult, E, Barbault, F, Paquet, F, Huynh-Dinh, T, Paoletti, J, Genest, D, Lancelot, G. | | Deposit date: | 2001-08-21 | | Release date: | 2002-12-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | HIV-1(Lai) genomic RNA: combined used of NMR and molecular dynamics simulation for studying
the structure and internal dynamics of a mutated SL1 hairpin.
EUR.BIOPHYS.J., 31, 2002
|
|
1T8V
 
 | |
