3BG0
 
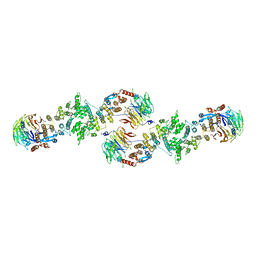 | | Architecture of a Coat for the Nuclear Pore Membrane | | Descriptor: | Nucleoporin NUP145, Protein SEC13 homolog | | Authors: | Hoelz, A. | | Deposit date: | 2007-11-23 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Architecture of a coat for the nuclear pore membrane.
Cell(Cambridge,Mass.), 131, 2007
|
|
3B37
 
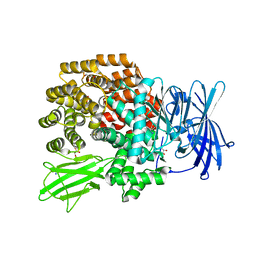 | |
1ZFB
 
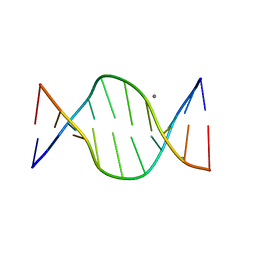 | | GGC Duplex B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*CP*CP*GP*GP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-20 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3B34
 
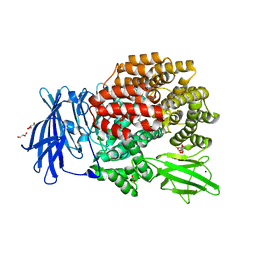 | |
3B9M
 
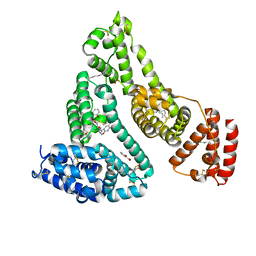 | | Human serum albumin complexed with myristate, 3'-azido-3'-deoxythymidine (AZT) and salicylic acid | | Descriptor: | 2-HYDROXYBENZOIC ACID, 3'-azido-3'-deoxythymidine, MYRISTIC ACID, ... | | Authors: | Zhu, L, Yang, F, Chen, L, Meehan, E.J, Huang, M. | | Deposit date: | 2007-11-05 | | Release date: | 2008-05-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A new drug binding subsite on human serum albumin and drug-drug interaction studied by X-ray crystallography
J.Struct.Biol., 162, 2008
|
|
1ZF0
 
 | | B-DNA | | Descriptor: | 5'-D(*CP*CP*GP*TP*TP*AP*AP*CP*GP*G)-3', CALCIUM ION | | Authors: | Hays, F.A, Teegarden, A.T, Jones, Z.J.R, Harms, M, Raup, D, Watson, J, Cavaliere, E, Ho, P.S. | | Deposit date: | 2005-04-19 | | Release date: | 2005-05-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | How sequence defines structure: a crystallographic map of DNA structure and conformation.
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3BIN
 
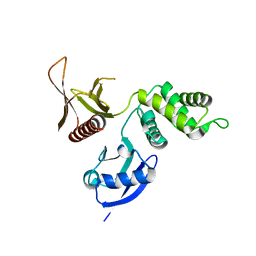 | | Structure of the DAL-1 and TSLC1 (372-383) complex | | Descriptor: | Band 4.1-like protein 3, Cell adhesion molecule 1 | | Authors: | Busam, R.D, Arrowsmith, C.H, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Sagemark, J, Svensson, L, Thorsell, A.G, Tresaugues, L, Van Den Berg, S, Weigelt, J, Welin, M, Berglund, H, Persson, C, Hallberg, B.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of tumor suppressor in lung cancer 1 (TSLC1) binding to differentially expressed in adenocarcinoma of the lung (DAL-1/4.1B)
J.Biol.Chem., 286, 2011
|
|
3B0P
 
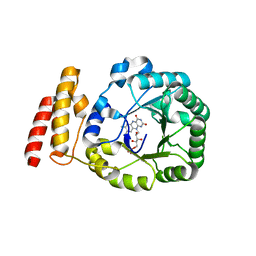 | | tRNA-dihydrouridine synthase from Thermus thermophilus | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase | | Authors: | Yu, F, Tanaka, Y, Yamashita, K, Nakamura, A, Yao, M, Tanaka, I. | | Deposit date: | 2011-06-12 | | Release date: | 2011-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of dihydrouridine formation on tRNA
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3B2Z
 
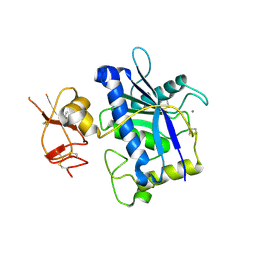 | | Crystal Structure of ADAMTS4 (apo form) | | Descriptor: | ADAMTS-4, CALCIUM ION, ZINC ION | | Authors: | Mosyak, L, Stahl, M, Somers, W. | | Deposit date: | 2007-10-19 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of the two major aggrecan degrading enzymes, ADAMTS4 and ADAMTS5.
Protein Sci., 17, 2008
|
|
3B60
 
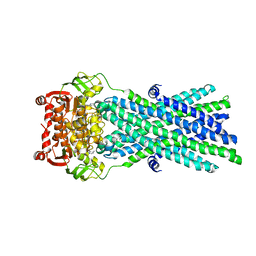 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP, higher resolution form | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1ZTV
 
 | |
1ZUP
 
 | |
1ZUD
 
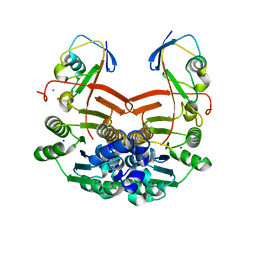 | | Structure of ThiS-ThiF protein complex | | Descriptor: | Adenylyltransferase thiF, CALCIUM ION, SODIUM ION, ... | | Authors: | Ealick, S.E, Lehmann, C. | | Deposit date: | 2005-05-30 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of the Escherichia coli ThiS-ThiF Complex, a Key Component of the Sulfur Transfer System in Thiamin Biosynthesis.
Biochemistry, 45, 2006
|
|
1ZT4
 
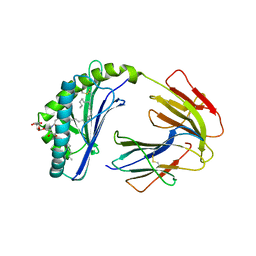 | | The crystal structure of human CD1d with and without alpha-Galactosylceramide | | Descriptor: | Beta-2-microglobulin, N-{(1S,2R,3S)-1-[(ALPHA-D-GALACTOPYRANOSYLOXY)METHYL]-2,3-DIHYDROXYHEPTADECYL}HEXACOSANAMIDE, T-cell surface glycoprotein CD1d | | Authors: | Koch, M, Stronge, V.S, Shepherd, D, Gadola, S.D, Mathew, B, Ritter, G, Fersht, A.R, Besra, G.S, Schmidt, R.R, Jones, E.Y, Cerundolo, V, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2005-05-26 | | Release date: | 2005-07-19 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of human CD1d with and without alpha-galactosylceramide
Nat.Immunol., 6, 2005
|
|
3B44
 
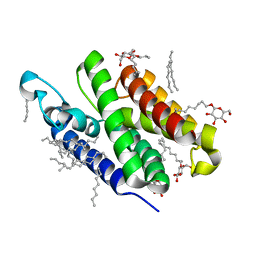 | | Crystal structure of GlpG W136A mutant | | Descriptor: | glpG, nonyl beta-D-glucopyranoside | | Authors: | Wang, Y, Maegawa, S, Akiyama, Y, Ha, Y. | | Deposit date: | 2007-10-23 | | Release date: | 2008-01-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG.
J.Mol.Biol., 374, 2007
|
|
3BAP
 
 | |
3BG9
 
 | |
3BH4
 
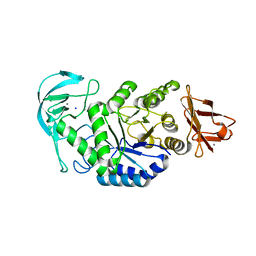 | | High resolution crystal structure of Bacillus amyloliquefaciens alpha-amylase | | Descriptor: | Alpha-amylase, CALCIUM ION, SODIUM ION | | Authors: | Alikhajeh, J, Khajeh, K, Ranjbar, B, Naderi-Manesh, H, Lin, Y.H, Liu, M.Y, Chen, C.J. | | Deposit date: | 2007-11-28 | | Release date: | 2008-12-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of Bacillus amyloliquefaciens alpha-amylase at high resolution: implications for thermal stability.
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3B0J
 
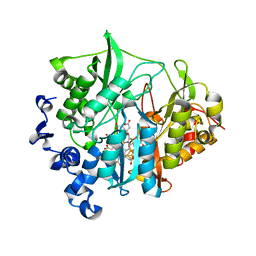 | | M175E mutant of assimilatory nitrite reductase (Nii3) from tobbaco leaf | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-10 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3B5Y
 
 | | Crystal Structure of MsbA from Salmonella typhimurium with AMPPNP | | Descriptor: | Lipid A export ATP-binding/permease protein msbA, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Ward, A, Reyes, C.L, Yu, J, Roth, C.B, Chang, G. | | Deposit date: | 2007-10-26 | | Release date: | 2007-12-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Flexibility in the ABC transporter MsbA: Alternating access with a twist.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B0H
 
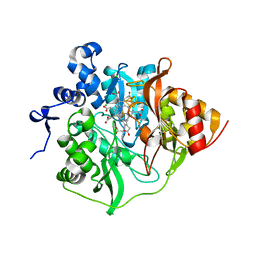 | | Assimilatory nitrite reductase (Nii4) from tobbaco root | | Descriptor: | CHLORIDE ION, IRON/SULFUR CLUSTER, Nitrite reductase, ... | | Authors: | Nakano, S, Takahashi, M, Sakamoto, A, Morikawa, H, Katayanagi, K. | | Deposit date: | 2011-06-09 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.306 Å) | | Cite: | Structure-function relationship of assimilatory nitrite reductases from the leaf and root of tobacco based on high resolution structures
Protein Sci., 21, 2012
|
|
3BCY
 
 | | Crystal structure of YER067W | | Descriptor: | Protein YER067W | | Authors: | Kozlov, G, Gehring, K. | | Deposit date: | 2007-11-13 | | Release date: | 2008-11-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional study of YER067W, a new protein involved in yeast metabolism control and drug resistance.
Plos One, 5, 2010
|
|
3C1U
 
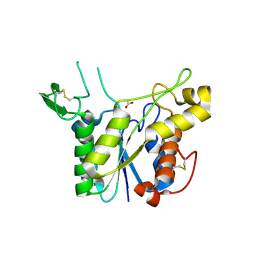 | | D192N mutant of Rhamnogalacturonan acetylesterase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Rhamnogalacturonan acetylesterase | | Authors: | Langkilde, A, Lo Leggio, L, Navarro Poulsen, J.C, Molgaard, A, Larsen, S. | | Deposit date: | 2008-01-24 | | Release date: | 2008-08-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Short strong hydrogen bonds in proteins: a case study of rhamnogalacturonan acetylesterase
ACTA CRYSTALLOGR.,SECT.D, 64, 2008
|
|
3C2J
 
 | |
3BK2
 
 | | Crystal Structure Analysis of the RNase J/UMP complex | | Descriptor: | GLYCEROL, Metal dependent hydrolase, SULFATE ION, ... | | Authors: | de la Sierra-Gallay, I.L, Zig, L, Putzer, H. | | Deposit date: | 2007-12-05 | | Release date: | 2008-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the dual activity of RNase J
Nat.Struct.Mol.Biol., 15, 2008
|
|
