1DJB
 
 | |
7UJQ
 
 | | Cocrystal structure of human CaMKII-alpha (CAMK2A)kinase domain and GluN2B | | Descriptor: | Calcium/calmodulin-dependent protein kinase type II subunit alpha, GLYCEROL, Glutamate receptor ionotropic, ... | | Authors: | Ozden, C, Foster, J.C, Stratton, M.M, Garman, S.C. | | Deposit date: | 2022-03-31 | | Release date: | 2022-04-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | CaMKII binds both substrates and activators at the active site.
Cell Rep, 40, 2022
|
|
1DOK
 
 | | MONOCYTE CHEMOATTRACTANT PROTEIN 1, P-FORM | | Descriptor: | MONOCYTE CHEMOATTRACTANT PROTEIN 1, SULFATE ION | | Authors: | Lubkowski, J, Bujacz, G, Boque, L, Wlodawer, A, Domaille, P.J, Handel, T.M. | | Deposit date: | 1996-11-27 | | Release date: | 1997-03-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The structure of MCP-1 in two crystal forms provides a rare example of variable quaternary interactions.
Nat.Struct.Biol., 4, 1997
|
|
1DQK
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE IN THE REDUCED STATE AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | FE (II) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
1E0P
 
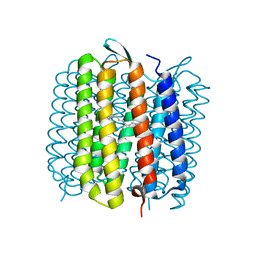 | | L intermediate of bacteriorhodopsin | | Descriptor: | BACTERIORHODOPSIN, GROUND STATE, RETINAL | | Authors: | Royant, A, Edman, K, Ursby, T, Pebay-Peyroula, E, Landau, E.M, Neutze, R. | | Deposit date: | 2000-04-04 | | Release date: | 2000-08-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Helix Deformation is Coupled to Vectorial Proton Transport in Bacteriorhodopsin'S Photocycle
Nature, 406, 2000
|
|
5A7B
 
 | |
1DQI
 
 | | CRYSTAL STRUCTURE OF SUPEROXIDE REDUCTASE FROM P. FURIOSUS IN THE OXIDIZED STATE AT 1.7 ANGSTROMS RESOLUTION | | Descriptor: | FE (III) ION, SUPEROXIDE REDUCTASE | | Authors: | Yeh, A.P, Hu, Y, Jenney Jr, F.E, Adams, M.W.W, Rees, D.C. | | Deposit date: | 2000-01-04 | | Release date: | 2000-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of the superoxide reductase from Pyrococcus furiosus in the oxidized and reduced states.
Biochemistry, 39, 2000
|
|
1DV6
 
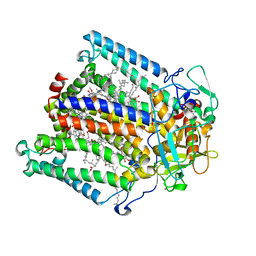 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE WITH THE PROTON TRANSFER INHIBITOR ZN2+ | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CHLORIDE ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2000-01-19 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the binding sites of the proton transfer inhibitors Cd2+ and Zn2+ in bacterial reaction centers.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
7M1R
 
 | |
1DS8
 
 | | PHOTOSYNTHETIC REACTION CENTER FROM RHODOBACTER SPHAEROIDES IN THE CHARGE-NEUTRAL DQAQB STATE WITH THE PROTON TRANSFER INHIBITOR CD2+ | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, CADMIUM ION, ... | | Authors: | Axelrod, H.L, Abresch, E.C, Paddock, M.L, Okamura, M.Y, Feher, G. | | Deposit date: | 2000-01-07 | | Release date: | 2000-03-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Determination of the binding sites of the proton transfer inhibitors Cd2+ and Zn2+ in bacterial reaction centers.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1DTN
 
 | |
1DJA
 
 | |
1E2N
 
 | | HPT + HMTT | | Descriptor: | 6-{[4-(HYDROXYMETHYL)-5-METHYL-2,6-DIOXOHEXAHYDROPYRIMIDIN-5-YL]METHYL}-5-METHYLPYRIMIDINE-2,4(1H,3H)-DIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Vogt, J, Scapozza, L, Schulz, G.E. | | Deposit date: | 2000-05-23 | | Release date: | 2001-03-31 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
1DZN
 
 | | Asp170Ser mutant of vanillyl-alcohol oxidase | | Descriptor: | 2-methoxy-4-[(1E)-prop-1-en-1-yl]phenol, FLAVIN-ADENINE DINUCLEOTIDE, VANILLYL-ALCOHOL OXIDASE | | Authors: | Van Den heuvel, R.H.H, Fraaije, M.W, Van Berkel, W.J.H, Mattevi, A. | | Deposit date: | 2000-03-05 | | Release date: | 2000-03-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Asp170 is Crucial for the Redox Properties of Vanillyl-Alcohol Oxidase
J.Biol.Chem., 275, 2000
|
|
1E2P
 
 | | Thymidine kinase, DHBT | | Descriptor: | 6-[3-HYDROXY-2-(HYDROXYMETHYL)PROPYL]-5-METHYL-2,4(1H,3H)-PYRIMIDINEDIONE, SULFATE ION, THYMIDINE KINASE | | Authors: | Schulz, G.E, Kessler, U. | | Deposit date: | 2000-05-23 | | Release date: | 2001-04-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Effect of Substrate Binding on the Conformation and Structural Stability of Herpes Simplex Virus Type 1 Thymidine Kinase
Protein Sci., 10, 2001
|
|
7SZK
 
 | | Cryo-EM structure of 27a bound to E. coli RNAP and rrnBP1 promoter complex | | Descriptor: | (2S,7R,7aR,13aP,16Z,18E,20S,21S,22R,23R,24R,25S,26R,27S,28E)-5,21,23-trihydroxy-27-methoxy-2,4,16,20,22,24,26-heptamethyl-10-[4-(2-methylpropyl)piperazin-1-yl]-12-({4-[(morpholin-4-yl)methyl]phenyl}methoxy)-1,6,15-trioxo-1,2,7,7a-tetrahydro-6H-2,7-(epoxypentadeca[1,11,13]trienoimino)[1]benzofuro[4,5-a]phenoxazin-25-yl acetate, DNA (5'-D(P*CP*TP*CP*GP*TP*AP*GP*AP*GP*TP*CP*CP*GP*TP*GP*TP*CP*A)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Shin, Y, Murakami, K.S. | | Deposit date: | 2021-11-28 | | Release date: | 2022-07-13 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Optimization of Benzoxazinorifamycins to Improve Mycobacterium tuberculosis RNA Polymerase Inhibition and Treatment of Tuberculosis.
Acs Infect Dis., 8, 2022
|
|
7SZJ
 
 | |
5SY4
 
 | |
5SY9
 
 | |
7NEN
 
 | |
7UE8
 
 | | PANK3 complex structure with compound PZ-3890 | | Descriptor: | 1,2-ETHANEDIOL, 1-[4-(6-chloropyridazin-3-yl)piperazin-1-yl]-2-(4-cyclopropyl-3-fluorophenyl)ethan-1-one, ACETATE ION, ... | | Authors: | White, S.W, Yun, M, Lee, R.E. | | Deposit date: | 2022-03-21 | | Release date: | 2023-03-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Relief of CoA sequestration and restoration of mitochondrial function in a mouse model of propionic acidemia.
J Inherit Metab Dis, 46, 2023
|
|
7L9X
 
 | | Structure of VPS4B in complex with an allele-specific covalent inhibitor | | Descriptor: | N-{3-[(8-phenyl[1,2,4]triazolo[1,5-a]pyridin-2-yl)amino]phenyl}propanamide, SULFATE ION, Vacuolar protein sorting-associated protein 4B | | Authors: | Grasso, M, Cupido, T, Kapoor, T.M. | | Deposit date: | 2021-01-05 | | Release date: | 2021-04-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | A chemical genetics approach to examine the functions of AAA proteins.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7N4N
 
 | | BACE-2 in complex with ligand 36 | | Descriptor: | 1,2-ETHANEDIOL, Beta-secretase 2, N-{3-[(2S,5R)-6-amino-2-(fluoromethyl)-5-(methanesulfonyl)-5-methyl-2,3,4,5-tetrahydropyridin-2-yl]-4-fluorophenyl}-6-methoxypyrimidine-4-carboxamide, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-04 | | Release date: | 2021-10-06 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
7N66
 
 | | BACE-1 in complex with ligand 12 | | Descriptor: | Beta-secretase 1, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2021-06-07 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | JNJ-67569762, A 2-Aminotetrahydropyridine-Based Selective BACE1 Inhibitor Targeting the S3 Pocket: From Discovery to Clinical Candidate.
J.Med.Chem., 64, 2021
|
|
5URC
 
 | | Design, Synthesis, Functional and Biological Evaluation of Ether and Ester Derivatives of the Antisickling Agent 5-HMF for the Treatment of Sickle Cell Disease | | Descriptor: | (5-formylfuran-2-yl)methyl acetate, 5-HYDROXYMETHYL-FURFURAL, CARBON MONOXIDE, ... | | Authors: | Pagare, P.P, Safo, R.S, Gazi, A. | | Deposit date: | 2017-02-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Ester and Ether Derivatives of Antisickling Agent 5-HMF for the Treatment of Sickle Cell Disease.
Mol. Pharm., 14, 2017
|
|
