5JFU
 
 | | HIV-1 wild Type protease with GRL-007-14A (a Adamantane P1-Ligand with bis-THF in P2 and benzylamine in P1') | | Descriptor: | (3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl {(2S,3R)-4-{benzyl[(4-methoxyphenyl)sulfonyl]amino}-3-hydroxy-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, Protease | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
1BP5
 
 | | HUMAN SERUM TRANSFERRIN, RECOMBINANT N-TERMINAL LOBE, APO FORM | | Descriptor: | PROTEIN (SERUM TRANSFERRIN) | | Authors: | Jeffrey, P.D, Bewley, M.C, Macgillivray, R.T.A, Mason, A.B, Woodworth, R.C, Baker, E.N. | | Deposit date: | 1998-08-12 | | Release date: | 1999-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ligand-induced conformational change in transferrins: crystal structure of the open form of the N-terminal half-molecule of human transferrin.
Biochemistry, 37, 1998
|
|
1IE8
 
 | | Crystal Structure Of The Nuclear Receptor For Vitamin D Ligand Binding Domain Bound to KH1060 | | Descriptor: | 5-(2-{1-[1-(4-ETHYL-4-HYDROXY-HEXYLOXY)-ETHYL]-7A-METHYL-OCTAHYDRO-INDEN-4-YLIDENE}-ETHYLIDENE)-4-METHYLENE-CYCLOHEXANE-1,3-DIOL, VITAMIN D3 RECEPTOR | | Authors: | Tocchini-Valentini, G, Rochel, N, Wurtz, J.M, Mitschler, A, Moras, D. | | Deposit date: | 2001-04-09 | | Release date: | 2001-05-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
5JG1
 
 | | HIV-1 wild Type protease with GRL-031-14A (a Adamantane P1-Ligand with tetrahydropyrano-tetrahydrofuran in P2 and isobutylamine in P1') | | Descriptor: | (3R,3aS,7aR)-hexahydro-4H-furo[2,3-b]pyran-3-yl {(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-[(3R,5R,7R)-tricyclo[3.3.1.1~3,7~]decan-1-yl]butan-2-yl}carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2016-04-19 | | Release date: | 2016-09-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Probing Lipophilic Adamantyl Group as the P1-Ligand for HIV-1 Protease Inhibitors: Design, Synthesis, Protein X-ray Structural Studies, and Biological Evaluation.
J.Med.Chem., 59, 2016
|
|
9F5N
 
 | |
9F5O
 
 | |
5UGM
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with Edaglitazone | | Descriptor: | (5R)-5-({4-[2-(5-methyl-2-phenyl-1,3-oxazol-4-yl)ethoxy]-1-benzothiophen-7-yl}methyl)-1,3-thiazolidine-2,4-dione, Peroxisome proliferator-activated receptor gamma, nonanoic acid | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2017-01-09 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cooperative cobinding of synthetic and natural ligands to the nuclear receptor PPAR gamma.
Elife, 7, 2018
|
|
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | Descriptor: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
5NVY
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-acetamidopropanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl) pyrrolidine-2-carboxamide (ligand 11) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-acetamidopropanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Soares, P, Gadd, M.S, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
8BJA
 
 | |
5NVW
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 6) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclopropylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5NVX
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(1-fluorocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 10) | | Descriptor: | Elongin-B, Elongin-C, N-[(1-fluorocyclopropyl)carbonyl]-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5NW2
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-3,3-dimethyl-2-(oxetane-3-carboxamido)butanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 19) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(oxetan-3-ylcarbonylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
5UPS
 
 | | Acyl-CoA synthetase PtmA2 from Streptomyces platensis in complex with SBNP663 ligand | | Descriptor: | 5'-O-[(R)-hydroxy{[(7beta,8alpha,9beta,10alpha,11beta,13alpha)-7-hydroxy-19-oxo-11,16-epoxykauran-19-yl]oxy}phosphoryl]adenosine, Acyl-CoA synthetase PtmA2, FORMIC ACID, ... | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Endres, M, Babnigg, G, Rudolf, J.D, Chang, C.-Y, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2017-02-03 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Natural separation of the acyl-CoA ligase reaction results in a non-adenylating enzyme.
Nat. Chem. Biol., 14, 2018
|
|
4W9J
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-((S)-2-acetamido-4-methylpentanamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 13) | | Descriptor: | N-acetyl-L-leucyl-3-methyl-L-valyl-(4R)-4-hydroxy-N-[4-(4-methyl-1,3-thiazol-5-yl)benzyl]-L-prolinamide, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2, ... | | Authors: | Gadd, M.S, Galdeano, C, van Molle, I, Ciulli, A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Guided Design and Optimization of Small Molecules Targeting the Protein-Protein Interaction between the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase and the Hypoxia Inducible Factor (HIF) Alpha Subunit with in Vitro Nanomolar Affinities.
J.Med.Chem., 57, 2014
|
|
3ST5
 
 | | Crystal structure of wild-type HIV-1 protease with C3-Substituted Hexahydrocyclopentafuranyl Urethane as P2-Ligand, GRL-0489A | | Descriptor: | (3R,3aR,5R,6aR)-3-hydroxyhexahydro-2H-cyclopenta[b]furan-5-yl [(2S,3R)-3-hydroxy-4-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}-1-phenylbutan-2-yl]carbamate, CHLORIDE ION, Protease | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Design of HIV-1 Protease Inhibitors with C3-Substituted Hexahydrocyclopentafuranyl Urethanes as P2-Ligands: Synthesis, Biological Evaluation, and Protein-Ligand X-ray Crystal Structure.
J.Med.Chem., 54, 2011
|
|
6FT5
 
 | | Structure of A3_A3, an artificial bi-domain protein based on two identical alphaRep A3 domains | | Descriptor: | GLYCEROL, SULFATE ION, alphaRep A3_A3 | | Authors: | Li de la Sierra-Gallay, I, Leger, C, Di Meo, T. | | Deposit date: | 2018-02-20 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Ligand-induced conformational switch in an artificial bidomain protein scaffold.
Sci Rep, 9, 2019
|
|
5NVV
 
 | | pVHL:EloB:EloC in complex with (2S,4R)-4-hydroxy-1-((S)-2-(2-hydroxyacetamido)-3,3-dimethylbutanoyl)-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 3) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-3,3-dimethyl-2-(2-oxidanylethanoylamino)butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Galdeano, C, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
6FSQ
 
 | |
4S14
 
 | | Crystal structure of the orphan nuclear receptor RORgamma ligand-binding domain in complex with 4alpha-caboxyl, 4beta-methyl-zymosterol (4ACD8) | | Descriptor: | (3beta,4alpha,5beta,14beta)-3-hydroxy-4-methylcholesta-8,24-diene-4-carboxylic acid, Nuclear receptor ROR-gamma, Nuclear receptor-interacting protein 1 | | Authors: | Huang, P, Santori, F.R, Littman, D.R, Rastinejad, F. | | Deposit date: | 2015-01-07 | | Release date: | 2015-02-11 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.542 Å) | | Cite: | Identification of Natural ROR gamma Ligands that Regulate the Development of Lymphoid Cells.
Cell Metab, 21, 2015
|
|
5NW1
 
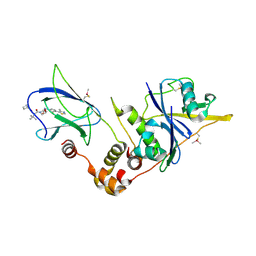 | | pVHL:EloB:EloC in complex with (2S,4R)-1-((S)-2-(cyclobutanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (ligand 18) | | Descriptor: | (2~{S},4~{R})-1-[(2~{S})-2-(cyclobutylcarbonylamino)-3,3-dimethyl-butanoyl]-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Soares, P, Ciulli, A. | | Deposit date: | 2017-05-04 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Group-Based Optimization of Potent and Cell-Active Inhibitors of the von Hippel-Lindau (VHL) E3 Ubiquitin Ligase: Structure-Activity Relationships Leading to the Chemical Probe (2S,4R)-1-((S)-2-(1-Cyanocyclopropanecarboxamido)-3,3-dimethylbutanoyl)-4-hydroxy-N-(4-(4-methylthiazol-5-yl)benzyl)pyrrolidine-2-carboxamide (VH298).
J. Med. Chem., 61, 2018
|
|
8W1V
 
 | | The beta2 adrenergic receptor bound to a bitopic ligand | | Descriptor: | (2S)-1-[(3-{1-[4-(4-{(2S)-2-hydroxy-3-[(propan-2-yl)amino]propoxy}phenyl)butyl]-1H-1,2,3-triazol-4-yl}propyl)amino]-3-(2-propylphenoxy)propan-2-ol, Beta-2 adrenergic receptor,Endolysin, Lauryl Maltose Neopentyl Glycol, ... | | Authors: | Gaiser, B, Danielsen, M, Xu, X, Jorgensen, K, Fronik, P, Marcher-Rorsted, E, Wrobe, T, Hirata, K, Liu, X, Mathiesen, J, Pedersen, D. | | Deposit date: | 2024-02-19 | | Release date: | 2024-07-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bitopic Ligands Support the Presence of a Metastable Binding Site at the beta 2 Adrenergic Receptor.
J.Med.Chem., 67, 2024
|
|
4O3A
 
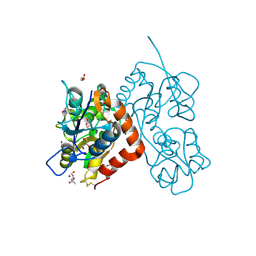 | | Crystal structure of the glua2 ligand-binding domain in complex with L-aspartate at 1.80 a resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, F, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3B
 
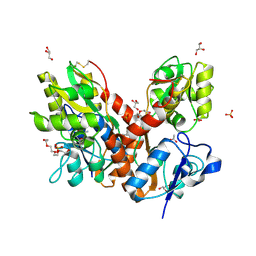 | | Crystal structure of an open/closed glua2 ligand-binding domain dimer at 1.91 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Krintel, C, de Rabassa, A.C, Frydenvang, K, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.906 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
4O3C
 
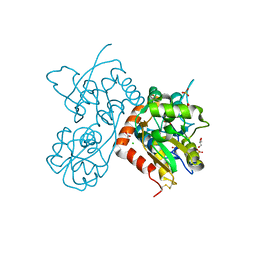 | | Crystal structure of the GLUA2 ligand-binding domain in complex with L-aspartate at 1.50 A resolution | | Descriptor: | ACETATE ION, ASPARTIC ACID, CHLORIDE ION, ... | | Authors: | Krintel, C, Frydenvang, K, Kaern, A.M, Gajhede, M, Kastrup, J.S. | | Deposit date: | 2013-12-18 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | L-Asp is a useful tool in the purification of the ionotropic glutamate receptor A2 ligand-binding domain.
Febs J., 281, 2014
|
|
