1SUA
 
 | | SUBTILISIN BPN' | | Descriptor: | SUBTILISIN BPN', TETRAPEPTIDE ALA-LEU-ALA-LEU | | Authors: | Almog, O, Gilliland, G.L. | | Deposit date: | 1997-01-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of calcium-independent subtilisin BPN' with restored thermal stability folded without the prodomain.
Proteins, 31, 1998
|
|
1SUB
 
 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
1SUC
 
 | |
1SUD
 
 | | CALCIUM-INDEPENDENT SUBTILISIN BY DESIGN | | Descriptor: | ACETONE, CALCIUM ION, POTASSIUM ION, ... | | Authors: | Gallagher, T, Bryan, P, Gilliland, G.L. | | Deposit date: | 1992-06-10 | | Release date: | 1994-01-31 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Calcium-independent subtilisin by design.
Proteins, 16, 1993
|
|
1SUE
 
 | | SUBTILISIN BPN' FROM BACILLUS AMYLOLIQUEFACIENS, MUTANT | | Descriptor: | DIISOPROPYL PHOSPHONATE, SODIUM ION, SUBTILISIN BPN' | | Authors: | Gallagher, D.T, Bryan, P, Pan, Q, Gilliland, G.L. | | Deposit date: | 1998-02-17 | | Release date: | 1998-10-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Mechanism of ionic strength dependence of crystal growth rates in a subtilisin variant.
J.Cryst.Growth, 193, 1998
|
|
1SUF
 
 | | Carbon Monoxide Dehydrogenase from Carboxydothermus hydrogenoformans-Inactive state | | Descriptor: | Carbon Monoxide Dehydrogenase 2, FE(4)-NI(1)-S(5) CLUSTER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Dobbek, H, Svetlitchnyi, V, Liss, J, Meyer, O. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Carbon Monoxide Induced Decomposition of the Active Site [Ni-4Fe-5S] Cluster of CO Dehydrogenase
J.Am.Chem.Soc., 126, 2004
|
|
1SUG
 
 | | 1.95 A structure of apo protein tyrosine phosphatase 1B | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Protein-tyrosine phosphatase, ... | | Authors: | Pedersen, A.K, Peters, G.H, Moller, K.B, Iversen, L.F, Kastrup, J.S. | | Deposit date: | 2004-03-26 | | Release date: | 2004-09-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Water-molecule network and active-site flexibility of apo protein tyrosine phosphatase 1B.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1SUH
 
 | | AMINO-TERMINAL DOMAIN OF EPITHELIAL CADHERIN IN THE CALCIUM BOUND STATE, NMR, 20 STRUCTURES | | Descriptor: | EPITHELIAL CADHERIN | | Authors: | Overduin, M, Tong, K.I, Kay, C.M, Ikura, M. | | Deposit date: | 1996-01-30 | | Release date: | 1996-07-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N and 13C resonance assignments and monomeric structure of the amino-terminal extracellular domain of epithelial cadherin.
J.Biomol.NMR, 7, 1996
|
|
1SUI
 
 | | Alfalfa caffeoyl coenzyme A 3-O-methyltransferase | | Descriptor: | CALCIUM ION, Caffeoyl-CoA O-methyltransferase, FERULOYL COENZYME A, ... | | Authors: | Ferrer, J.-L, Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Alfalfa Caffeoyl Coenzyme A 3-O-Methyltransferase
Plant Physiol., 137, 2005
|
|
1SUJ
 
 | |
1SUL
 
 | | Crystal Structure of the apo-YsxC | | Descriptor: | GTP-binding protein YsxC | | Authors: | Ruzheinikov, S.N, Das, K.S, Sedelnikova, S.E, Baker, P.J, Artymiuk, P.J, Garcia-Lara, J, Foster, S.J, Rice, D.W. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the Open and Closed Conformations of the GTP-binding Protein YsxC from Bacillus subtilis.
J.Mol.Biol., 339, 2004
|
|
1SUM
 
 | | Crystal structure of a hypothetical protein at 2.0 A resolution | | Descriptor: | CALCIUM ION, FE (III) ION, NICKEL (II) ION, ... | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a PhoU protein homologue: a new class of metalloprotein containing multinuclear iron clusters.
J.Biol.Chem., 280, 2005
|
|
1SUO
 
 | | Structure of mammalian cytochrome P450 2B4 with bound 4-(4-chlorophenyl)imidazole | | Descriptor: | 4-(4-CHLOROPHENYL)IMIDAZOLE, Cytochrome P450 2B4, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Scott, E.E, White, M.A, He, Y.A, Johnson, E.F, Stout, C.D, Halpert, J.R. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mammalian cytochrome P450 2B4 complexed with 4-(4-chlorophenyl)imidazole at 1.9 {angstrom} resolution: Insight into the range of P450 conformations and coordination of redox partner binding.
J.Biol.Chem., 279, 2004
|
|
1SUP
 
 | | SUBTILISIN BPN' AT 1.6 ANGSTROMS RESOLUTION: ANALYSIS OF DISCRETE DISORDER AND COMPARISON OF CRYSTAL FORMS | | Descriptor: | CALCIUM ION, SODIUM ION, SUBTILISIN BPN', ... | | Authors: | Gallagher, D.T, Oliver, J.D, Betzel, C, Gilliland, G.L. | | Deposit date: | 1995-08-14 | | Release date: | 1995-11-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Subtilisin BPN' at 1.6 A resolution: analysis for discrete disorder and comparison of crystal forms.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1SUQ
 
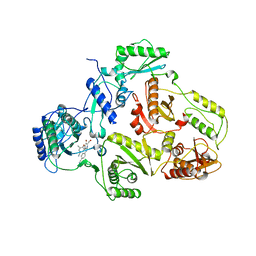 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R185545 | | Descriptor: | (6-[4-(AMINOMETHYL)-2,6-DIMETHYLPHENOXY]-2-{[4-(AMINOMETHYL)PHENYL]AMINO}-5-BROMOPYRIMIDIN-4-YL)METHANOL, MAGNESIUM ION, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-26 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
1SUR
 
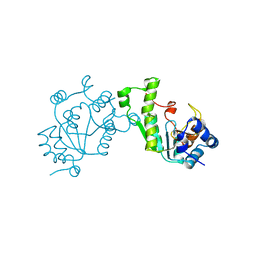 | | PHOSPHO-ADENYLYL-SULFATE REDUCTASE | | Descriptor: | PAPS REDUCTASE | | Authors: | Sinning, I, Savage, H. | | Deposit date: | 1998-04-01 | | Release date: | 1999-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of phosphoadenylyl sulphate (PAPS) reductase: a new family of adenine nucleotide alpha hydrolases.
Structure, 5, 1997
|
|
1SUS
 
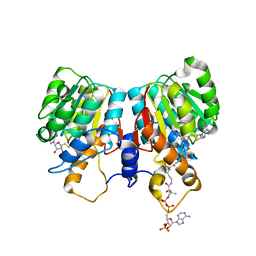 | | Crystal structure of alfalfa feruoyl coenzyme A 3-O-methyltransferase | | Descriptor: | CALCIUM ION, Caffeoyl-CoA O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Ferrer, J.-L, Zubieta, C, Dixon, R.A, Noel, J.P. | | Deposit date: | 2004-03-26 | | Release date: | 2005-03-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of Alfalfa Caffeoyl Coenzyme A 3-O-Methyltransferase
Plant Physiol., 137, 2005
|
|
1SUT
 
 | |
1SUU
 
 | |
1SUV
 
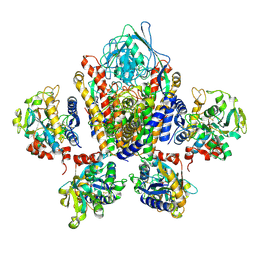 | | Structure of Human Transferrin Receptor-Transferrin Complex | | Descriptor: | CARBONATE ION, FE (III) ION, Serotransferrin, ... | | Authors: | Cheng, Y, Zak, O, Aisen, P, Harrison, S.C, Walz, T. | | Deposit date: | 2004-03-26 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | Structure of the Human Transferrin Receptor-Transferrin Complex
Cell(Cambridge,Mass.), 116, 2004
|
|
1SUW
 
 | | Crystal structure of a NAD kinase from Archaeoglobus fulgidus in complex with its substrate and product: Insights into the catalysis of NAD kinase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Probable inorganic polyphosphate/ATP-NAD kinase | | Authors: | Liu, J, Lou, Y, Yokota, H, Adams, P.D, Kim, R, Kim, S.-H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal Structures of an NAD Kinase from Archaeoglobus fulgidus in Complex with ATP, NAD, or NADP
J.Mol.Biol., 354, 2005
|
|
1SUX
 
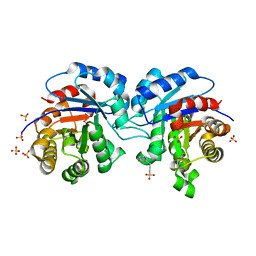 | | CRYSTALLOGRAPHIC ANALYSIS OF THE COMPLEX BETWEEN TRIOSEPHOSPHATE ISOMERASE FROM TRYPANOSOMA CRUZI AND 3-(2-benzothiazolylthio)-1-propanesulfonic acid | | Descriptor: | 3-(2-BENZOTHIAZOLYLTHIO)-1-PROPANESULFONIC ACID, SULFATE ION, Triosephosphate isomerase, ... | | Authors: | Tellez-Valencia, A, Olivares-Illana, V, Hernandez-Santoyo, A, Perez-Montfort, R, Costas, M, Rodriguez-Romero, A, Tuena De Gomez-Puyou, M, Gomez-Puyou, A. | | Deposit date: | 2004-03-26 | | Release date: | 2004-08-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inactivation of triosephosphate isomerase from Trypanosoma cruzi by an agent that perturbs its dimer interface.
J.Mol.Biol., 341, 2004
|
|
1SUY
 
 | |
1SUZ
 
 | |
1SV0
 
 | | Crystal Structure Of Yan-SAM/Mae-SAM Complex | | Descriptor: | Ets DNA-binding protein pokkuri, modulator of the activity of Ets CG15085-PA | | Authors: | Qiao, F, Song, H, Kim, C.A, Sawaya, M.R, Hunter, J.B, Gingery, M, Rebay, I, Courey, A.J, Bowie, J.U. | | Deposit date: | 2004-03-26 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Derepression by depolymerization; structural insights into the regulation of yan by mae.
Cell(Cambridge,Mass.), 118, 2004
|
|
