1ZBI
 
 | | Bacillus halodurans RNase H catalytic domain mutant D132N in complex with 12-mer RNA/DNA hybrid | | Descriptor: | 5'-D(*GP*AP*AP*TP*CP*AP*GP*GP*TP*GP*TP*C)-3', 5'-R(*GP*AP*CP*AP*CP*CP*UP*GP*AP*UP*UP*C)-3', MAGNESIUM ION, ... | | Authors: | Nowotny, M, Gaidamakov, S.A, Crouch, R.J, Yang, W. | | Deposit date: | 2005-04-08 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structures of RNase H Bound to an RNA/DNA Hybrid: Substrate Specificity and Metal-Dependent Catalysis.
Cell(Cambridge,Mass.), 121, 2005
|
|
1AOC
 
 | | JAPANESE HORSESHOE CRAB COAGULOGEN | | Descriptor: | COAGULOGEN, SULFATE ION | | Authors: | Bergner, A, Oganessyan, V, Muta, T, Iwanaga, S, Typke, D, Huber, R, Bode, W. | | Deposit date: | 1996-11-28 | | Release date: | 1997-04-21 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a coagulogen, the clotting protein from horseshoe crab: a structural homologue of nerve growth factor.
EMBO J., 15, 1996
|
|
2CFQ
 
 | | Sugar Free Lactose Permease at neutral pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
2CI3
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase crystal form I | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2CK2
 
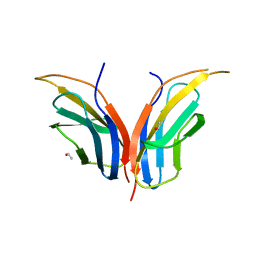 | | Structure of core-swapped mutant of fibronectin | | Descriptor: | ACETYL GROUP, HUMAN FIBRONECTIN | | Authors: | Ng, S.P, Billings, K.S, Ohashi, T, Allen, M.D, Best, R.B, Randles, L.G, Erickson, H.P, Clarke, J. | | Deposit date: | 2006-04-10 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designing an Extracellular Matrix Protein with Enhanced Mechanical Stability
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
1Z1W
 
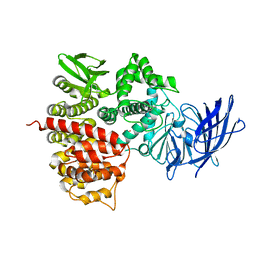 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | Descriptor: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | Authors: | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-05-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
2CCZ
 
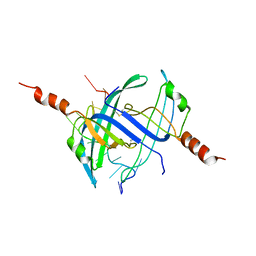 | | Crystal structure of E. coli primosomol protein PriB bound to ssDNA | | Descriptor: | 5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP *TP*TP*TP*TP*T)-3', PRIMOSOMAL REPLICATION PROTEIN N | | Authors: | Huang, C.-Y, Hsu, C.-H, Wu, H.-N, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2006-01-19 | | Release date: | 2006-09-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Complexed Crystal Structure of Replication Restart Primsome Protein Prib Reveals a Novel Single-Stranded DNA-Binding Mode.
Nucleic Acids Res., 34, 2006
|
|
1Z34
 
 | | Crystal structure of Trichomonas vaginalis purine nucleoside phosphorylase complexed with 2-fluoro-2'-deoxyadenosine | | Descriptor: | 5-(6-AMINO-2-FLUORO-PURIN-9-YL)-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-OL, purine nucleoside phosphorylase | | Authors: | Zhang, Y, Wang, W.H, Wu, S.W, Wang, C.C, Ealick, S.E. | | Deposit date: | 2005-03-10 | | Release date: | 2005-03-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of a subversive substrate of Trichomonas vaginalis purine nucleoside phosphorylase and the crystal structure of the enzyme-substrate complex.
J.Biol.Chem., 280, 2005
|
|
1UZP
 
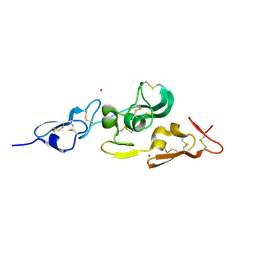 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, Sm bound form cbEGF23 domain only. | | Descriptor: | FIBRILLIN-1, SAMARIUM (III) ION | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-15 | | Release date: | 2004-04-08 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
2CI4
 
 | | Crystal Structure of Dimethylarginine dimethylaminohydrolase I crystal form II | | Descriptor: | NG, NG-DIMETHYLARGININE DIMETHYLAMINOHYDROLASE 1 | | Authors: | Frey, D, Braun, O, Briand, C, Vasak, M, Grutter, M.G. | | Deposit date: | 2006-03-17 | | Release date: | 2006-05-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the Mammalian Nos Regulator Dimethylarginine Dimethylaminohydrolase: A Basis for the Design of Specific Inhibitors.
Structure, 14, 2006
|
|
2C78
 
 | | EF-Tu complexed with a GTP analog and the antibiotic pulvomycin | | Descriptor: | (1S,2S,3E,5E,7E,10S,11S,12S)-12-[(2R,4E,6E,8Z,10R,12E,14E,16Z,18S,19Z)-10,18-DIHYDROXY-12,16,19-TRIMETHYL-11,22-DIOXOOX ACYCLODOCOSA-4,6,8,12,14,16,19-HEPTAEN-2-YL]-2,11-DIHYDROXY-1,10-DIMETHYL-9-OXOTRIDECA-3,5,7-TRIEN-1-YL 6-DEOXY-2,4-DI-O-METHYL-BETA-L-GALACTOPYRANOSIDE, ELONGATION FACTOR TU-A, MAGNESIUM ION, ... | | Authors: | Parmeggiani, A, Krab, I.M, Okamura, S, Nielsen, R.C, Nyborg, J, Nissen, P. | | Deposit date: | 2005-11-18 | | Release date: | 2006-03-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis of the action of pulvomycin and GE2270 A on elongation factor Tu.
Biochemistry, 45, 2006
|
|
2CTV
 
 | |
1ZHW
 
 | | Structure of yeast oxysterol binding protein Osh4 in complex with 20-hydroxycholesterol | | Descriptor: | 20-HYDROXYCHOLESTEROL, KES1 protein, LEAD (II) ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-26 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
2CHR
 
 | |
2CND
 
 | | STRUCTURAL STUDIES ON CORN NITRATE REDUCTASE: REFINED STRUCTURE OF THE CYTOCHROME B REDUCTASE FRAGMENT AT 2.5 ANGSTROMS, ITS ADP COMPLEX AND AN ACTIVE SITE MUTANT AND MODELING OF THE CYTOCHROME B DOMAIN | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-DEPENDENT NITRATE REDUCTASE | | Authors: | Lu, G, Lindqvist, Y, Schneider, G. | | Deposit date: | 1995-02-01 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural studies on corn nitrate reductase: refined structure of the cytochrome b reductase fragment at 2.5 A, its ADP complex and an active-site mutant and modeling of the cytochrome b domain.
J.Mol.Biol., 248, 1995
|
|
1ZI7
 
 | | Structure of truncated yeast oxysterol binding protein Osh4 | | Descriptor: | KES1 protein, SULFATE ION | | Authors: | Im, Y.J, Raychaudhuri, S, Prinz, W.A, Hurley, J.H. | | Deposit date: | 2005-04-27 | | Release date: | 2005-09-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural mechanism for sterol sensing and transport by OSBP-related proteins
Nature, 437, 2005
|
|
2CSM
 
 | |
1UXY
 
 | | MURB MUTANT WITH SER 229 REPLACED BY ALA, COMPLEX WITH ENOLPYRUVYL-UDP-N-ACETYLGLUCOSAMINE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, URIDINE DIPHOSPHO-N-ACETYLENOLPYRUVYLGLUCOSAMINE REDUCTASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL) BUTYRIC ACID | | Authors: | Benson, T.E, Walsh, C.T, Hogle, J.M. | | Deposit date: | 1996-11-08 | | Release date: | 1997-04-01 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structures of the S229A mutant and wild-type MurB in the presence of the substrate enolpyruvyl-UDP-N-acetylglucosamine at 1.8-A resolution.
Biochemistry, 36, 1997
|
|
1UWB
 
 | | TYR 181 CYS HIV-1 RT/8-CL TIBO | | Descriptor: | 5-CHLORO-8-METHYL-7-(3-METHYL-BUT-2-ENYL)-6,7,8,9-TETRAHYDRO-2H-2,7,9A-TRIAZA-BENZO[CD]AZULENE-1-THIONE, REVERSE TRANSCRIPTASE | | Authors: | Das, K, Ding, J, Hsiou, Y, Arnold, E. | | Deposit date: | 1996-11-21 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structures of 8-Cl and 9-Cl TIBO complexed with wild-type HIV-1 RT and 8-Cl TIBO complexed with the Tyr181Cys HIV-1 RT drug-resistant mutant.
J.Mol.Biol., 264, 1996
|
|
2CSN
 
 | | BINARY COMPLEX OF CASEIN KINASE-1 WITH CKI7 | | Descriptor: | CASEIN KINASE-1, N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, SULFATE ION | | Authors: | Xu, R.-M, Cheng, X. | | Deposit date: | 1995-10-11 | | Release date: | 1996-03-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for selectivity of the isoquinoline sulfonamide family of protein kinase inhibitors.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
2CAH
 
 | |
1URT
 
 | | MURINE CARBONIC ANHYDRASE V | | Descriptor: | CARBONIC ANHYDRASE V, ZINC ION | | Authors: | Boriack-Sjodin, P.A, Christianson, D.W. | | Deposit date: | 1996-07-03 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure-based design of an intramolecular proton transfer site in murine carbonic anhydrase V.
Biochemistry, 35, 1996
|
|
2CBL
 
 | | N-TERMINAL DOMAIN OF CBL IN COMPLEX WITH ITS BINDING SITE ON ZAP-70 | | Descriptor: | CALCIUM ION, PROTO-ONCOGENE CBL, ZAP-70 | | Authors: | Meng, W, Sawasdikosol, S, Burakoff, S.J, Eck, M.J. | | Deposit date: | 1998-08-28 | | Release date: | 1999-05-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the amino-terminal domain of Cbl complexed to its binding site on ZAP-70 kinase.
Nature, 398, 1999
|
|
1AHR
 
 | |
1V6T
 
 | |
