4QR4
 
 | | Brd4 Bromodomain 1 complex with its novel inhibitors | | Descriptor: | 2-chloro-N-cyclopentyl-5-(2-oxo-2,3-dihydro-1,3-thiazol-4-yl)benzenesulfonamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Cao, D.Y, Chen, T.T, Xu, Y.C. | | Deposit date: | 2014-06-30 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fragment-based drug discovery of 2-thiazolidinones as BRD4 inhibitors: 2. Structure-based optimization
J.Med.Chem., 58, 2015
|
|
9QLE
 
 | | Human myoferlin (1-1997) in complex with an MSP2N2 lipid nanodisc (15 mol% DOPS, 2 mol% PI(4,5)P2) | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, CALCIUM ION, Myoferlin | | Authors: | Cretu, C, Moser, T. | | Deposit date: | 2025-03-20 | | Release date: | 2025-06-04 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights into lipid membrane binding by human ferlins.
Embo J., 44, 2025
|
|
9UL1
 
 | | Crystal structure of Tibetan wild boar SLA-1*Z0301 for 2.32 angstrom | | Descriptor: | ALA-LEU-LEU-SER-SER-LYS-THR-SER-VAL, Beta-2-microglobulin, MHC class I antigen | | Authors: | Fan, S, Wang, Y. | | Deposit date: | 2025-04-18 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.323 Å) | | Cite: | Structural basis of Tibetan wild boar SLA-1*Z0301 reveals conserved peptide presentation and potential high-altitude adaptation.
Int.J.Biol.Macromol., 320, 2025
|
|
8JXS
 
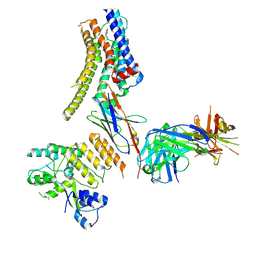 | | Structure of nanobody-bound DRD1_PF-6142 complex | | Descriptor: | 4-[3-methyl-4-(6-methylimidazo[1,2-a]pyrazin-5-yl)phenoxy]furo[3,2-c]pyridine, D(1A) dopamine receptor, Fab 8D3 heavy chain, ... | | Authors: | Zhuang, Y, Xu, Y, Fan, L, Wang, S, Xu, H.E. | | Deposit date: | 2023-07-01 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of psychedelic LSD recognition at dopamine D 1 receptor.
Neuron, 112, 2024
|
|
9QKV
 
 | |
9OWD
 
 | | Human WRN helicase in complex with allosteric ligand Compound 9 | | Descriptor: | 1,2-ETHANEDIOL, Bifunctional 3'-5' exonuclease/ATP-dependent helicase WRN, DIMETHYL SULFOXIDE, ... | | Authors: | Palte, R.L, Koglin, M, Maskos, K, Tauchert, M.J. | | Deposit date: | 2025-06-02 | | Release date: | 2025-09-10 | | Last modified: | 2025-09-17 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | High-throughput evaluation of novel WRN inhibitors.
Slas Discov, 35, 2025
|
|
9UDF
 
 | | Cryo-EM structure of Na+-translocating NADH-ubiquinone oxidoreductase NqrB-G141A mutant from Vibrio cholerae reduced by NADH, with bound korormicin A, shifted state | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ishikawa-Fukuda, M, Kishikawa, J, Kato, T, Murai, M. | | Deposit date: | 2025-04-06 | | Release date: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (2.93 Å) | | Cite: | The Na + -pumping mechanism driven by redox reactions in the NADH-quinone oxidoreductase from Vibrio cholerae relies on dynamic conformational changes.
Biorxiv, 2025
|
|
9MTU
 
 | |
9NXH
 
 | | An alpha-l-arabinofuranosidase (AtAbf43C) from Acetivibrio thermocellus DSM1313 bound to arabinofuranose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 43, MAGNESIUM ION, ... | | Authors: | Galindo, J.L, Jeffrey, P.D, Conway, J.M. | | Deposit date: | 2025-03-25 | | Release date: | 2025-08-20 | | Last modified: | 2025-08-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Functional and structural characterization of AtAbf43C: an exo-1,5-alpha-L-arabinofuranosidase from Acetivibrio thermocellus DSM1313.
Biochem.J., 482, 2025
|
|
9SOS
 
 | |
9OU1
 
 | | Crystal structure of maize AKR4C13 in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, Aldose reductase, AKR4C13, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R, Sousa, S.M. | | Deposit date: | 2025-05-28 | | Release date: | 2025-07-02 | | Last modified: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Functional genomics and structural insights into maize aldo-keto reductase-4 family: Stress metabolism and substrate specificity in embryos.
J.Biol.Chem., 301, 2025
|
|
9QJE
 
 | | USP7 Covalently Bound to N-(6-Fluoro-3-nitropyridin-2-yl)-5-(1-methyl-1H-pyrazol-4-yl)isoquinolin-3-amine (GCL36, 7a) with Partial Occupancy | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Stahlecker, J, Ernst, L.N, Gehringer, M, Stehle, T, Boeckler, F.M. | | Deposit date: | 2025-03-19 | | Release date: | 2025-09-10 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Design, Synthesis, and Molecular Evaluation of S N Ar-Reactive N-(6-Fluoro-3-Nitropyridin-2-yl)Isoquinolin-3-Amines as Covalent USP7 Inhibitors Reveals an Unconventional Binding Mode.
Arch Pharm, 358, 2025
|
|
9OMS
 
 | | X-ray structure of human acetylcholinesterase (hAChE) in complex with bis-oxime reactivator LG-1922 | | Descriptor: | 2-(hydroxyimino)-N-{2-[(3S)-1-(3-{[(2E)-2-(hydroxyimino)acetyl]amino}propyl)piperidin-3-yl]ethyl}acetamide, Acetylcholinesterase, GLYCEROL, ... | | Authors: | Kovalevsky, A, Radic, Z, Gerlits, O. | | Deposit date: | 2025-05-14 | | Release date: | 2025-07-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Kinetic and structural evidence for specific DMSO interference with reversible binding of uncharged bis-oximes to hAChE and their reactivation kinetics of OP-hAChE.
Chem.Biol.Interact., 419, 2025
|
|
4WBP
 
 | |
7PEB
 
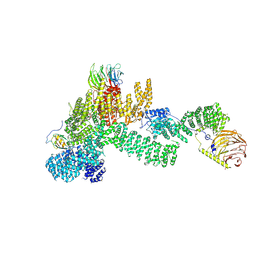 | |
7PEC
 
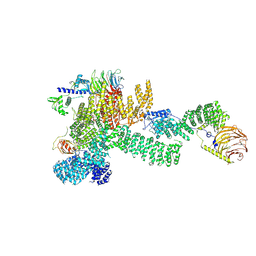 | |
7PEA
 
 | |
9RWD
 
 | | High-resolution structure of human SHMT2 with covalently bound PLP (internal aldimine) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Warlich, A, Ruszkowski, M, Nawrot, D. | | Deposit date: | 2025-07-09 | | Release date: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-resolution structure of human SHMT2 with covalently bound PLP (internal aldimine)
To Be Published
|
|
8HAS
 
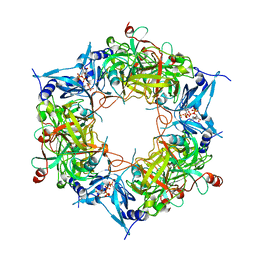 | | NARROW LEAF 1-close from Japonica | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein NARROW LEAF 1 | | Authors: | Zhang, S.J, He, Y.J, Wang, N, Zhang, W.J, Liu, C.M. | | Deposit date: | 2022-10-26 | | Release date: | 2024-05-01 | | Last modified: | 2025-07-16 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | NARROW LEAF 1-close from Japonica
To Be Published
|
|
8OV3
 
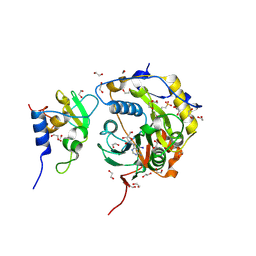 | | SARS-CoV-2 nsp10-16 methyltransferase in complex with 5-Iodotubercidin | | Descriptor: | (2R,3R,4S,5R)-2-(4-AMINO-5-IODO-7H-PYRROLO[2,3-D]PYRIMIDIN-7-YL)-5-(HYDROXYMETHYL)TETRAHYDROFURAN-3,4-DIOL, 1,2-ETHANEDIOL, 2'-O-methyltransferase nsp16, ... | | Authors: | Kremling, V, Sprenger, J, Oberthuer, D. | | Deposit date: | 2023-04-25 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 methyltransferase nsp10-16 in complex with natural and drug-like purine analogs for guiding structure-based drug discovery
Elife, 2024
|
|
8TUC
 
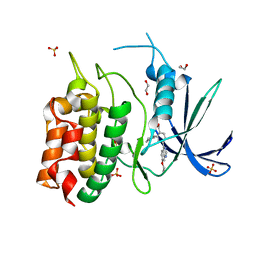 | | Unphosphorylated CaMKK2 in complex with CC-8977 | | Descriptor: | (4M)-2-cyclopentyl-4-(7-ethoxyquinazolin-4-yl)benzoic acid, 1,2-ETHANEDIOL, Calcium/calmodulin-dependent protein kinase kinase 2, ... | | Authors: | Bernard, S.M, Shanmugasundaram, V, D'Agostino, L. | | Deposit date: | 2023-08-16 | | Release date: | 2023-12-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Identification of Small Molecule Inhibitors and Ligand Directed Degraders of Calcium/Calmodulin Dependent Protein Kinase Kinase 1 and 2 (CaMKK1/2).
J.Med.Chem., 66, 2023
|
|
8OMS
 
 | |
7MAL
 
 | | HIV-1 Protease (I84V) in Complex with PU8 (LR4-06) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-[(2-ethylbutyl){[4-(hydroxymethyl)phenyl]sulfonyl}amino]-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | HIV-1 Protease (I84V) in Complex with PU8 (LR4-06)
To Be Published
|
|
8HAT
 
 | |
6VLG
 
 | | Crystal structure of mouse alpha 1,6-fucosyltransferase, FUT8 bound to GDP | | Descriptor: | Alpha-(1,6)-fucosyltransferase, GUANOSINE-5'-DIPHOSPHATE, SULFATE ION, ... | | Authors: | Jarva, M.A, Dramicanin, M, Lingford, J.P, Mao, R, John, A, Goddard-Borger, E.D. | | Deposit date: | 2020-01-23 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of substrate recognition and catalysis by fucosyltransferase 8.
J.Biol.Chem., 295, 2020
|
|
