6HTS
 
 | | Cryo-EM structure of the human INO80 complex bound to nucleosome | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin-related protein 5, Chromatin-remodeling ATPase INO80, ... | | Authors: | Ayala, R, Willhoft, O, Aramayo, R.J, Wilkinson, M, McCormack, E.A, Ocloo, L, Wigley, D.B, Zhang, X. | | Deposit date: | 2018-10-04 | | Release date: | 2018-11-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structure and regulation of the human INO80-nucleosome complex.
Nature, 556, 2018
|
|
5FZQ
 
 | | Designed TPR Protein M4N | | Descriptor: | DESIGNED TPR PROTEIN, SULFATE ION | | Authors: | Albrecht, R, Zhu, H, Hartmann, M.D. | | Deposit date: | 2016-03-15 | | Release date: | 2016-10-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.148 Å) | | Cite: | Origin of a folded repeat protein from an intrinsically disordered ancestor.
Elife, 5, 2016
|
|
2K8X
 
 | |
6E9F
 
 | | EsCas13d-crRNA-target RNA ternary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, RNA (27-MER), ... | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
6E9E
 
 | | EsCas13d-crRNA binary complex | | Descriptor: | EsCas13d, MAGNESIUM ION, crRNA (52-MER) | | Authors: | Zhang, C, Lyumkis, D. | | Deposit date: | 2018-08-01 | | Release date: | 2018-10-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural Basis for the RNA-Guided Ribonuclease Activity of CRISPR-Cas13d.
Cell, 175, 2018
|
|
5FZR
 
 | |
5FZS
 
 | |
5FZW
 
 | |
8J4I
 
 | | Unveiling the Role of Human Prohibitin 2 as a Mitochondrial Calcium Channel in Parkinson's Disease | | Descriptor: | Prohibitin-2 | | Authors: | Li, Y.Y, Fang, Y, Gao, Y.X, Ding, W, Yang, J, Liu, Y, Shen, B, Wang, J.F, Zhou, S. | | Deposit date: | 2023-04-20 | | Release date: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Human Prohibitin 2 is a Mitochondrial Ca2+-Selective Channel
To Be Published
|
|
6E8G
 
 | |
5F81
 
 | | Acoustic injectors for drop-on-demand serial femtosecond crystallography | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Roessler, C.G, Agarwal, R, Allaire, M, Alonso-Mori, R, Andi, B, Bachega, J.F.R, Bommer, M, Brewster, A.S, Browne, M.C, Chatterjee, R, Cho, E, Cohen, A.E, Cowan, M, Datwani, S, Davidson, V.L, Defever, J, Eaton, B, Ellson, R, Feng, Y, Ghislain, L.P, Glownia, J.M, Han, G, Hattne, J, Hellmich, J, Heroux, A, Ibrahim, M, Kern, J, Kuczewski, A, Lemke, H.T, Liu, P, Majlof, L, McClintock, W.M, Myers, S, Nelsen, S, Olechno, J, Orville, A.M, Sauter, N.K, Soares, A.S, Soltis, M.S, Song, H, Stearns, R.G, Tran, R, Tsai, Y, Uervirojnangkoorn, M, Wilmot, C.M, Yachandra, V, Yano, J, Yukl, E.T, Zhu, D, Zouni, A. | | Deposit date: | 2015-12-08 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Acoustic Injectors for Drop-On-Demand Serial Femtosecond Crystallography.
Structure, 24, 2016
|
|
6F0L
 
 | | S. cerevisiae MCM double hexamer bound to duplex DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (62-MER), DNA replication licensing factor MCM2, ... | | Authors: | Abid Ali, F, Pye, V.E, Douglas, M.E, Locke, J, Nans, A, Diffley, J.F.X, Costa, A. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-06 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (4.77 Å) | | Cite: | Cryo-EM structure of a licensed DNA replication origin.
Nat Commun, 8, 2017
|
|
6EZO
 
 | | Eukaryotic initiation factor EIF2B in complex with ISRIB | | Descriptor: | 2-(4-chloranylphenoxy)-~{N}-[4-[2-(4-chloranylphenoxy)ethanoylamino]cyclohexyl]ethanamide, Human eukaryotic initiation factor EIF2B epsilon subunits, Translation initiation factor eIF-2B subunit alpha, ... | | Authors: | Faille, A, Weis, F, Zyryanova, A, Warren, A.J, Ron, D. | | Deposit date: | 2017-11-16 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Binding of ISRIB reveals a regulatory site in the nucleotide exchange factor eIF2B.
Science, 359, 2018
|
|
7PGD
 
 | | PAF in 50 v/v % DMSO-water solution | | Descriptor: | Pc24g00380 protein | | Authors: | Czajlik, A, Batta, G. | | Deposit date: | 2021-08-13 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | DMSO-Induced Unfolding of the Antifungal Disulfide Protein PAF and Its Inactive Variant: A Combined NMR and DSC Study.
Int J Mol Sci, 24, 2023
|
|
2IQY
 
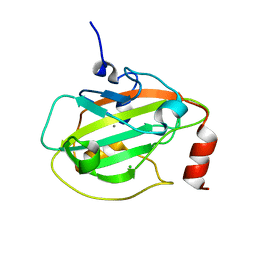 | | Rat Phosphatidylethanolamine-Binding Protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phosphatidylethanolamine-binding protein 1 | | Authors: | Kim, Y, Joachimiak, G, Heil, G.L, Koide, S, Joachimiak, A. | | Deposit date: | 2006-10-14 | | Release date: | 2007-09-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structure of Rat Phosphatidylethanolamine-Binding Protein
To be Published
|
|
4A3G
 
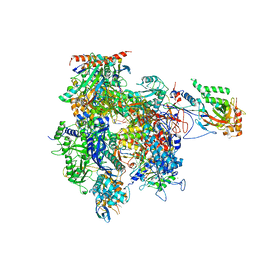 | | RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
6F38
 
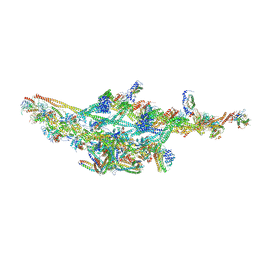 | | Cryo-EM structure of two dynein tail domains bound to dynactin and HOOK3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Urnavicius, L, Elshenawy, M.M, Morales-Rios, E, Motz, C, Yildiz, A, Carter, A.P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-01-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6.7 Å) | | Cite: | Cryo-EM shows how dynactin recruits two dyneins for faster movement.
Nature, 554, 2018
|
|
4A3J
 
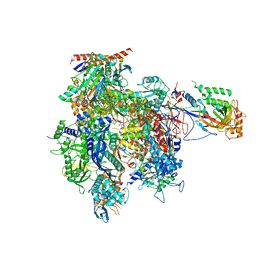 | | RNA Polymerase II initial transcribing complex with a 2nt DNA-RNA hybrid and soaked with GMPCPP | | Descriptor: | 5'-D(*AP*GP*CP*TP*AP*GP*CP*TP*TP*TP*CP*BRUP*AP*CP*CP *TP*GP*AP*AP*CP*AP*AP*CP*TP*AP*AP*CP)-3', 5'-D(*GP*TP*AP*GP*AP*AP*AP*GP*CP*TP*AP*GP*CP*TP)-3', 5'-R(*CP*AP)-3', ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Initial RNA Polymerase II Transcription.
Embo J., 30, 2011
|
|
6BBM
 
 | | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Replication protein P, Replicative DNA helicase | | Authors: | Chase, J, Catalano, A, Noble, A.J, Eng, E.T, Olinares, P.D.B, Molloy, K, Pakotiprapha, D, Samuels, M, Chain, B, des Georges, A, Jeruzalmi, D. | | Deposit date: | 2017-10-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanisms of opening and closing of the bacterial replicative helicase.
Elife, 7, 2018
|
|
6F3K
 
 | | Combined solid-state NMR, solution-state NMR and EM data for structure determination of the tetrahedral aminopeptidase TET2 from P. horikoshii | | Descriptor: | Tetrahedral aminopeptidase, ZINC ION | | Authors: | Gauto, D.F, Estrozi, L.F, Schwieters, C.D, Effantin, G, Macek, P, Sounier, R, Kerfah, R, Sivertsen, A.C, Colletier, J.P, Boisbouvier, J, Schoehn, G, Favier, A, Schanda, P. | | Deposit date: | 2017-11-28 | | Release date: | 2018-03-14 | | Last modified: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å), SOLID-STATE NMR, SOLUTION NMR | | Cite: | Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex.
Nat Commun, 10, 2019
|
|
5FR3
 
 | | X-ray crystal structure of aggregation-resistant protective antigen of Bacillus anthracis (mutant S559L T576E) | | Descriptor: | CALCIUM ION, GLYCEROL, PROTECTIVE ANTIGEN | | Authors: | Ganesan, A, Siekierska, A, Beerten, J, Brams, M, van Durme, J, De Baets, G, van der Kant, R, Gallardo, R, Ramakers, M, Langenberg, T, Wilkinson, H, De Smet, F, Ulens, C, Rousseau, F, Schymkowitz, J. | | Deposit date: | 2015-12-15 | | Release date: | 2016-01-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.935 Å) | | Cite: | Structural Hot Spots for the Solubility of Globular Proteins
Nat.Commun., 7, 2016
|
|
6FAY
 
 | | Teneurin3 monomer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Odz3 protein | | Authors: | Janssen, B.J.C, Meijer, D.H.M, van Bezouwen, L.S. | | Deposit date: | 2017-12-18 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of Teneurin adhesion receptors reveal an ancient fold for cell-cell interaction.
Nat Commun, 9, 2018
|
|
6H61
 
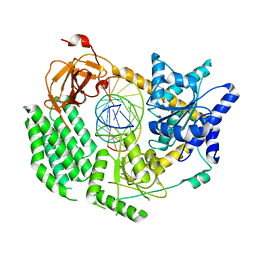 | | CryoEM structure of the MDA5-dsRNA filament with 89 degree twist and without nucleotide | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, RNA (5'-R(P*CP*GP*UP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*GP*A)-3'), RNA (5'-R(P*UP*CP*CP*AP*UP*GP*CP*GP*CP*AP*UP*GP*AP*CP*G)-3'), ... | | Authors: | Yu, Q, Qu, K, Modis, Y. | | Deposit date: | 2018-07-25 | | Release date: | 2018-11-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.02 Å) | | Cite: | Cryo-EM Structures of MDA5-dsRNA Filaments at Different Stages of ATP Hydrolysis.
Mol. Cell, 72, 2018
|
|
6FHL
 
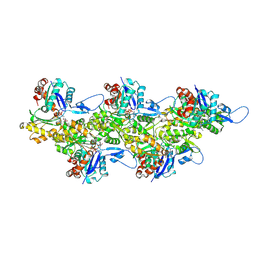 | | Cryo-EM structure of F-actin in complex with ADP-Pi | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Merino, F, Pospich, S, Funk, J, Wagner, T, Kuellmer, F, Arndt, H.-D, Bieling, P, Raunser, S. | | Deposit date: | 2018-01-15 | | Release date: | 2018-06-13 | | Last modified: | 2018-08-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural transitions of F-actin upon ATP hydrolysis at near-atomic resolution revealed by cryo-EM.
Nat. Struct. Mol. Biol., 25, 2018
|
|
4A3I
 
 | | RNA Polymerase II binary complex with DNA | | Descriptor: | 5'-D(*GP*GP*CP*AP*CP*AP*AP*CP*TP*GP*CP*GP*GP*CP*T)-3', DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, ... | | Authors: | Cheung, A.C.M, Sainsbury, S, Cramer, P. | | Deposit date: | 2011-09-30 | | Release date: | 2012-02-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis of initial RNA polymerase II transcription.
EMBO J., 30, 2011
|
|
