7OJP
 
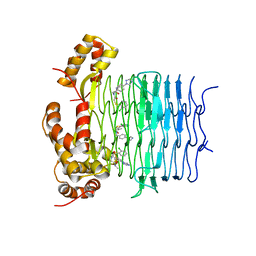 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 1 | | Descriptor: | 2-[2-(2-chlorophenyl)sulfanylethanoyl-[[4-(1,2,4-triazol-1-yl)phenyl]methyl]amino]-N-methyl-ethanamide, Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, MAGNESIUM ION | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
9O2R
 
 | |
4WDZ
 
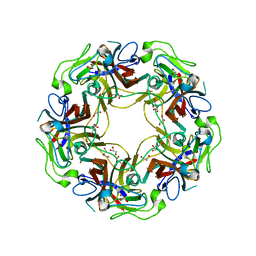 | | JC Polyomavirus VP1 five-fold pore mutant N221W | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Major capsid protein VP1 | | Authors: | Stroh, L.J, Stehle, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of a Pore in the Capsid of JC Polyomavirus Reduces Infectivity and Prevents Exposure of the Minor Capsid Proteins.
J.Virol., 89, 2015
|
|
6VD7
 
 | |
6MWR
 
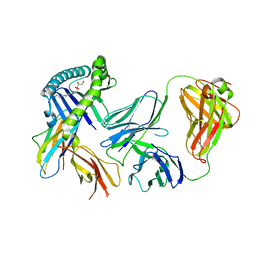 | | Recognition of MHC-like molecule | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, ... | | Authors: | Le Nours, J, Rossjohn, J. | | Deposit date: | 2018-10-30 | | Release date: | 2019-12-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A class of gamma delta T cell receptors recognize the underside of the antigen-presenting molecule MR1.
Science, 366, 2019
|
|
9S8Z
 
 | |
8CXU
 
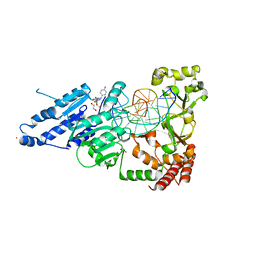 | | CamA Adenine Methyltransferase Complexed to Cognate Substrate DNA and Compound 2 | | Descriptor: | 1,2-ETHANEDIOL, DNA Strand 1, DNA Strand 2, ... | | Authors: | Horton, J.R, Zhou, J, Cheng, X. | | Deposit date: | 2022-05-22 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Systematic Design of Adenosine Analogs as Inhibitors of a Clostridioides difficile- Specific DNA Adenine Methyltransferase Required for Normal Sporulation and Persistence.
J.Med.Chem., 66, 2023
|
|
9S8V
 
 | |
8GI9
 
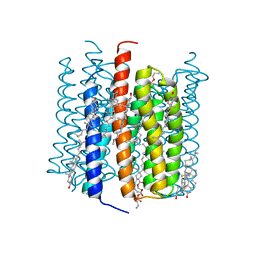 | | Cation channelrhodopsin from Hyphochytrium catenoides (HcCCR) embedded in peptidisc | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CHOLESTEROL, Cation Channelrhodopsin, ... | | Authors: | Morizumi, T, Kim, K, Li, H, Spudich, J.L, Ernst, O.P. | | Deposit date: | 2023-03-13 | | Release date: | 2023-07-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structures of channelrhodopsin paralogs in peptidiscs explain their contrasting K + and Na + selectivities.
Nat Commun, 14, 2023
|
|
8QEV
 
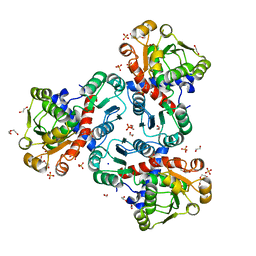 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with carbamoyl phosphate | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Ornithine transcarbamylase, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
6N2H
 
 | | Structure of D-ornithine/D-lysine decarboxylase from Salmonella typhimurium | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, D-ornithine/D-lysine decarboxylase, DIMETHYL SULFOXIDE | | Authors: | Phillips, R.S, Hoover, T.R. | | Deposit date: | 2018-11-13 | | Release date: | 2019-10-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal Structure of d-Ornithine/d-Lysine Decarboxylase, a Stereoinverting Decarboxylase: Implications for Substrate Specificity and Stereospecificity of Fold III Decarboxylases.
Biochemistry, 58, 2019
|
|
8QEU
 
 | | Crystal structure of ornithine transcarbamylase from Arabidopsis thaliana (AtOTC) in complex with ornithine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-ornithine, ... | | Authors: | Nielipinski, M, Pietrzyk-Brzezinska, A, Sekula, B. | | Deposit date: | 2023-09-01 | | Release date: | 2023-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural analysis and molecular substrate recognition properties of Arabidopsis thaliana ornithine transcarbamylase, the molecular target of phaseolotoxin produced by Pseudomonas syringae .
Front Plant Sci, 14, 2023
|
|
7O7J
 
 | | Crystal structure of the human HIPK3 kinase domain bound to abemaciclib | | Descriptor: | Homeodomain-interacting protein kinase 3, N-{5-[(4-ethylpiperazin-1-yl)methyl]pyridin-2-yl}-5-fluoro-4-[4-fluoro-2-methyl-1-(propan-2-yl)-1H-benzimidazol-6-yl]py rimidin-2-amine | | Authors: | Kaltheuner, I.H, Anand, K, Geyer, M. | | Deposit date: | 2021-04-13 | | Release date: | 2021-11-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Abemaciclib is a potent inhibitor of DYRK1A and HIP kinases involved in transcriptional regulation.
Nat Commun, 12, 2021
|
|
6PQK
 
 | |
6NCR
 
 | |
8CCD
 
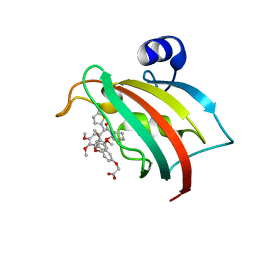 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)-2-(3,4,5-trimethoxyphenyl)ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCB
 
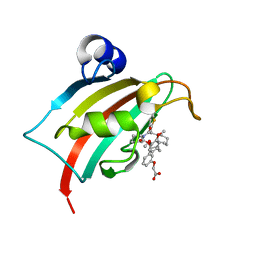 | | The Fk1 domain of FKBP51 in complex with 2-(3-((1R)-1-(((2S)-1-(2-(5-chlorothiophen-2-yl)-2-cyclohexylacetyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)-2-cyclohexyl-ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCE
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)butanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)butanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, GLYCEROL, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCC
 
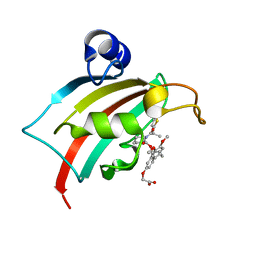 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)propanoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)propanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCF
 
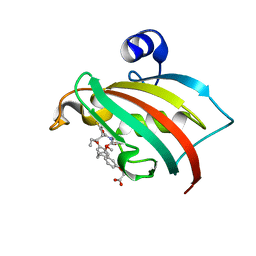 | | The Fk1 domain of FKBP51 in complex with 2-(3-((R)-1-(((S)-1-((S)-2-(5-chlorothiophen-2-yl)pent-4-enoyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-(5-chloranylthiophen-2-yl)pent-4-enoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
8CCH
 
 | | The Fk1 domain of FKBP51 in complex with 2-(3-((1R)-1-(((2S)-1-(2-cyclohexyl-2-(thiophen-2-yl)acetyl)piperidine-2-carbonyl)oxy)-3-(3,4-dimethoxyphenyl)propyl)phenoxy)acetic acid | | Descriptor: | 2-[3-[(1~{R})-1-[(2~{S})-1-[(2~{S})-2-cyclohexyl-2-thiophen-2-yl-ethanoyl]piperidin-2-yl]carbonyloxy-3-(3,4-dimethoxyphenyl)propyl]phenoxy]ethanoic acid, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Meyners, C, Knaup, F.H, Walz, C.M, Hausch, F. | | Deposit date: | 2023-01-27 | | Release date: | 2023-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structure-Based Discovery of a New Selectivity-Enabling Motif for the FK506-Binding Protein 51.
J.Med.Chem., 66, 2023
|
|
4WCO
 
 | | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A | | Descriptor: | ACETATE ION, C-type lectin domain family 2 member D, SULFATE ION, ... | | Authors: | Kita, S, Matsubara, H, Kasai, Y, Tamaoki, T, Okabe, Y, Fukuhara, H, Kamishikiryo, J, Ose, T, Kuroki, K, Maenaka, K. | | Deposit date: | 2014-09-05 | | Release date: | 2015-06-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of extracellular domain of human lectin-like transcript 1 (LLT1), the ligand for natural killer receptor-P1A
Eur.J.Immunol., 45, 2015
|
|
8T8N
 
 | |
8QDB
 
 | | Wdyg1p Ser256DHA (PSF) | | Descriptor: | 1,2-ETHANEDIOL, Yellowish-green 1-like protein | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
4UWJ
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a capped pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-N-(difluoromethyl)-4-[3-(piperidin-4-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
