1T3T
 
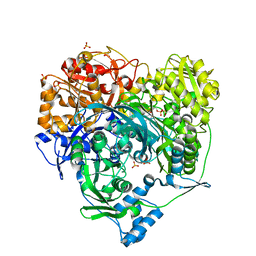 | | Structure of Formylglycinamide synthetase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Phosphoribosylformylglycinamidine synthase, ... | | Authors: | Ealick, S.E, Anand, R, Hoskin, A.A, Stubbe, J. | | Deposit date: | 2004-04-27 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Domain Organization of Salmonella typhimurium Formylglycinamide Ribonucleotide Amidotransferase Revealed by X-ray crystallography
Biochemistry, 43, 2004
|
|
1T4A
 
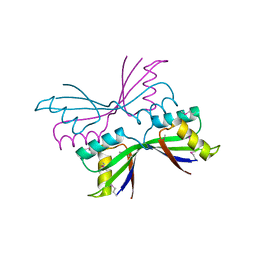 | | Structure of B. Subtilis PurS C2 Crystal Form | | Descriptor: | PurS | | Authors: | Anand, R, Hoskins, A.A, Bennett, E.M, Sintchak, M.D, Stubbe, J, Ealick, S.E. | | Deposit date: | 2004-04-28 | | Release date: | 2004-09-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A model for the Bacillus subtilis formylglycinamide ribonucleotide amidotransferase multiprotein complex.
Biochemistry, 43, 2004
|
|
7TCM
 
 | |
1T7L
 
 | |
1T4D
 
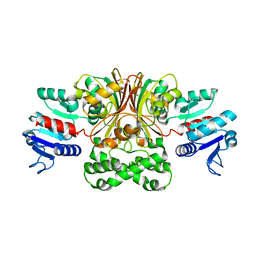 | | Crystal structure of Escherichia coli aspartate beta-semialdehyde dehydrogenase (EcASADH), at 1.95 Angstrom resolution | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
1TB4
 
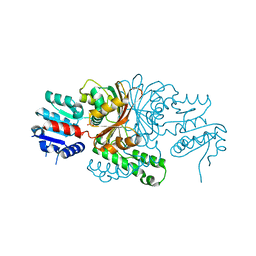 | |
1TEV
 
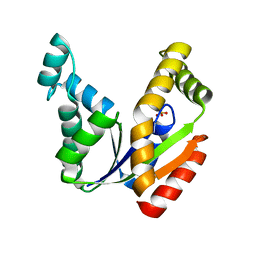 | | Crystal structure of the human UMP/CMP kinase in open conformation | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Segura-Pena, D, Sekulic, N, Ort, S, Konrad, M, Lavie, A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced Conformational Changes in Human UMP/CMP Kinase.
J.Biol.Chem., 279, 2004
|
|
1T4B
 
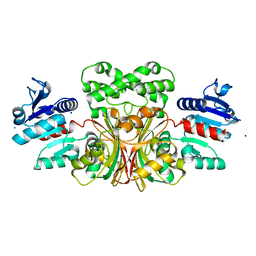 | | 1.6 Angstrom structure of Esherichia coli aspartate-semialdehyde dehydrogenase. | | Descriptor: | Aspartate-semialdehyde dehydrogenase, SODIUM ION | | Authors: | Nichols, C.E, Dhaliwal, B, Lockyer, M, Hawkins, A.R, Stammers, D.K. | | Deposit date: | 2004-04-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution Structures Reveal Details of Domain Closure and "Half-of-sites-reactivity" in Escherichia coli Aspartate beta-Semialdehyde Dehydrogenase.
J.Mol.Biol., 341, 2004
|
|
1T11
 
 | | Trigger Factor | | Descriptor: | Trigger factor | | Authors: | Ludlam, A.V, Moore, B.A, Xu, Z. | | Deposit date: | 2004-04-14 | | Release date: | 2004-09-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of ribosomal chaperone trigger factor from Vibrio cholerae.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6PZ1
 
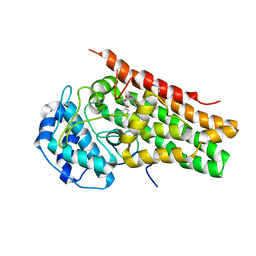 | | Crystal Structure of human Indoleamine 2,3-Dioxygenase 1 in complex with PF-06840003 in Active Site and Si site | | Descriptor: | (3R)-3-(5-fluoro-1H-indol-3-yl)pyrrolidine-2,5-dione, GLYCEROL, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Pham, K.N, Lewis-Ballester, A, Yeh, S.R. | | Deposit date: | 2019-07-31 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Basis of Inhibitor Selectivity in Human Indoleamine 2,3-Dioxygenase 1 and Tryptophan Dioxygenase.
J.Am.Chem.Soc., 141, 2019
|
|
1TA4
 
 | |
3EK6
 
 | |
7Z3N
 
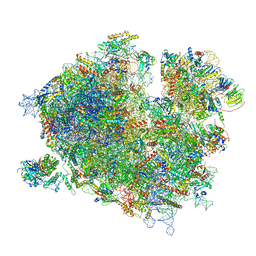 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-1 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7Z3O
 
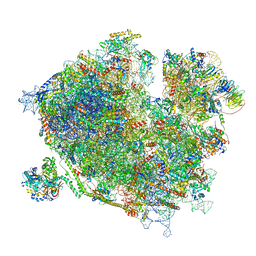 | | Cryo-EM structure of the ribosome-associated RAC complex on the 80S ribosome - RAC-2 conformation | | Descriptor: | 18S rRNA, 26S rRNA, 40S ribosomal protein S0, ... | | Authors: | Kisonaite, M, Wild, K, Sinning, I. | | Deposit date: | 2022-03-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural inventory of cotranslational protein folding by the eukaryotic RAC complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
5JQW
 
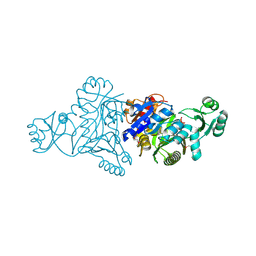 | | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP | | Descriptor: | ACETATE ION, ADENOSINE-5'-DIPHOSPHATE, N5-carboxyaminoimidazole ribonucleotide synthase | | Authors: | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-05 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The crystal structure of phosphoribosylaminoimidazole carboxylase ATPase subunit of Francisella tularensis subsp. tularensis SCHU S4 in complex with ADP
To Be Published
|
|
5K7X
 
 | |
5DJE
 
 | | Crystal structure of the zuotin homology domain (ZHD) from yeast Zuo1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shrestha, O.K, Bingman, C.A, Craig, E.A. | | Deposit date: | 2015-09-02 | | Release date: | 2016-09-28 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Dual interaction of the Hsp70 J-protein cochaperone Zuotin with the 40S and 60S ribosomal subunits.
Nat.Struct.Mol.Biol., 23, 2016
|
|
5CTJ
 
 | |
6VTY
 
 | |
6W1N
 
 | |
6W2J
 
 | | CPS1 bound to allosteric inhibitor H3B-374 | | Descriptor: | (2-fluoranyl-4-methoxy-phenyl)-[(3~{R},5~{R})-4-(2-fluoranyl-4-methoxy-phenyl)carbonyl-3,5-dimethyl-piperazin-1-yl]methanone, 1,2-ETHANEDIOL, Carbamoyl-phosphate synthase [ammonia], ... | | Authors: | Larsen, N.A, Nguyen, T.V. | | Deposit date: | 2020-03-05 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Discovery of 2,6-Dimethylpiperazines as Allosteric Inhibitors of CPS1.
Acs Med.Chem.Lett., 11, 2020
|
|
5VEV
 
 | |
3LOU
 
 | |
3MDO
 
 | |
9BEI
 
 | |
