6TY9
 
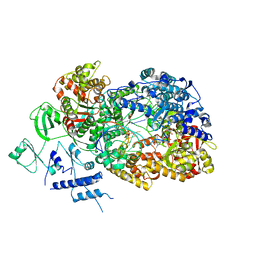 | | In situ structure of BmCPV RNA dependent RNA polymerase at initiation state | | Descriptor: | MAGNESIUM ION, Non-template RNA (5'-D(*(GTA))-R(P*GP*UP*AP*AP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*U)-3'), RNA-dependent RNA Polymerase, ... | | Authors: | Cui, Y.X, Zhang, Y.N, Sun, J.C, Zhou, Z.H. | | Deposit date: | 2019-08-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Conservative transcription in three steps visualized in a double-stranded RNA virus.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6U23
 
 | | EM structure of MPEG-1(w.t.) soluble pre-pore | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-19 | | Release date: | 2019-09-25 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
7OSR
 
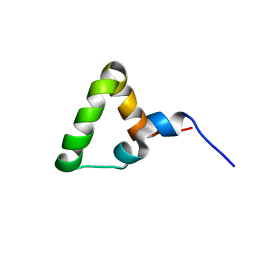 | |
7P2O
 
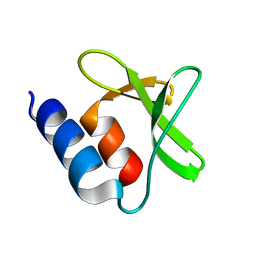 | |
6UGD
 
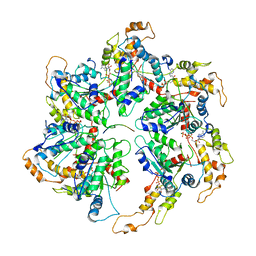 | |
6UHC
 
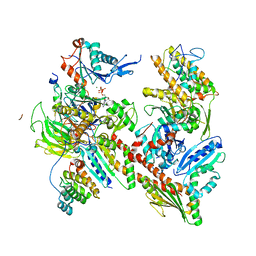 | | CryoEM structure of human Arp2/3 complex with bound NPFs | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | Zimmet, A, van Eeuwen, T, Dominguez, R. | | Deposit date: | 2019-09-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of NPF-bound human Arp2/3 complex and activation mechanism.
Sci Adv, 6, 2020
|
|
6UBO
 
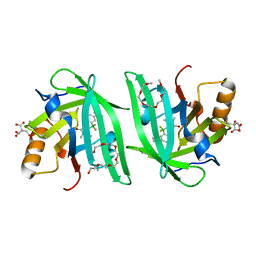 | | Fluorogen Activating Protein Dib1 | | Descriptor: | 12-(diethylamino)-2,2-bis(fluoranyl)-4,5-dimethyl-5-aza-3-azonia-2-boranuidatricyclo[7.4.0.0^{3,7}]trideca-1(13),3,7,9,11-pentaen-6-one, CITRIC ACID, Outer membrane lipoprotein Blc, ... | | Authors: | Muslinkina, L, Pletneva, N, Pletnev, V.Z, Pletnev, S. | | Deposit date: | 2019-09-12 | | Release date: | 2020-09-16 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure-Based Rational Design of Two Enhanced Bacterial Lipocalin Blc Tags for Protein-PAINT Super-resolution Microscopy.
Acs Chem.Biol., 15, 2020
|
|
7UBI
 
 | |
6TWE
 
 | |
8T3U
 
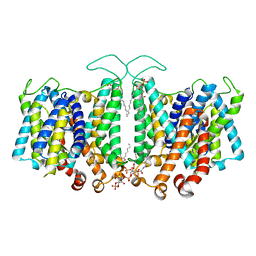 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
7PVB
 
 | |
8T44
 
 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CARBONATE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T45
 
 | |
6UGE
 
 | |
8T47
 
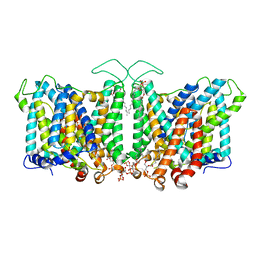 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaHCO3 Buffer: Form3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CARBONATE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-08 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
8T3R
 
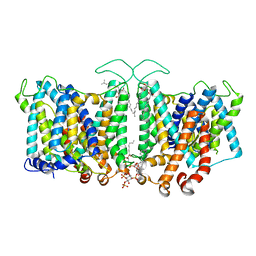 | | Cryo-EM Analysis of AE1 Structure in 100 mM NaCl Buffer: Form1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Band 3 anion transport protein, CHLORIDE ION, ... | | Authors: | Su, C.C. | | Deposit date: | 2023-06-07 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Cryo-EM structure of the band 3 anion transport protein
To Be Published
|
|
7UI5
 
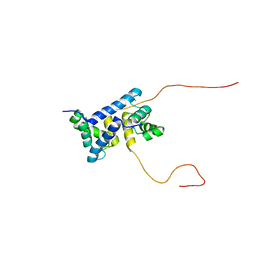 | | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9 | | Descriptor: | CALCIUM ION, Protein S100-A9 | | Authors: | Reardon, P.N, Harman, J.L, Costello, S.M, Warren, G.D, Phillips, S.R, Connor, P.J, Marqusee, S, Harms, M.J. | | Deposit date: | 2022-03-28 | | Release date: | 2022-10-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Evolution avoids a pathological stabilizing interaction in the immune protein S100A9.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7JOF
 
 | | Calcium-bound C2A Domain from Human Dysferlin | | Descriptor: | CALCIUM ION, Isoform 6 of Dysferlin | | Authors: | Tadayon, R, Wang, Y, Santamaria, L, Mercier, P, Forristal, C, Shaw, G.S. | | Deposit date: | 2020-08-06 | | Release date: | 2021-06-16 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Calcium binds and rigidifies the dysferlin C2A domain in a tightly coupled manner.
Biochem.J., 478, 2021
|
|
7UO6
 
 | |
8E53
 
 | | MicroED structure of proteinase K recorded on K3 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.7 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
6XOR
 
 | |
8E52
 
 | | MicroED structure of proteinase K recorded on K2 | | Descriptor: | CALCIUM ION, Proteinase K | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-11-13 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.8 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
8E54
 
 | | MicroED structure of triclinic lysozyme recorded on K3 | | Descriptor: | Lysozyme C, NITRATE ION | | Authors: | Clabbers, M.T.B, Martynowycz, M.W, Hattne, J, Nannenga, B.L, Gonen, T. | | Deposit date: | 2022-08-19 | | Release date: | 2022-09-21 | | Last modified: | 2024-10-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.2 Å) | | Cite: | Electron-counting MicroED data with the K2 and K3 direct electron detectors.
J.Struct.Biol., 214, 2022
|
|
6TZE
 
 | | Human CstF-64 RRM mutant - D50A | | Descriptor: | Cleavage stimulation factor subunit 2 | | Authors: | Latham, M.P, Masoumzadeh, E. | | Deposit date: | 2019-08-12 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A missense mutation in the CSTF2 gene that impairs the function of the RNA recognition motif and causes defects in 3' end processing is associated with intellectual disability in humans.
Nucleic Acids Res., 48, 2020
|
|
7URJ
 
 | |
