3NWV
 
 | | Human cytochrome c G41S | | Descriptor: | Cytochrome c, HEME C | | Authors: | Fagerlund, R.D, Wilbanks, S.M. | | Deposit date: | 2010-07-11 | | Release date: | 2011-03-09 | | Last modified: | 2019-10-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Proapoptotic G41S Mutation to Human Cytochrome c Alters the Heme Electronic Structure and Increases the Electron Self-Exchange Rate.
J.Am.Chem.Soc., 133, 2011
|
|
3NBT
 
 | | Crystal structure of trimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3NBS
 
 | | Crystal structure of dimeric cytochrome c from horse heart | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Taketa, M, Komori, H, Hirota, S, Higuchi, Y. | | Deposit date: | 2010-06-04 | | Release date: | 2010-07-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cytochrome c polymerization by successive domain swapping at the C-terminal helix
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3M97
 
 | | Structure of the soluble domain of cytochrome c552 with its flexible linker segment from Paracoccus denitrificans | | Descriptor: | Cytochrome c-552, HEME C, ZINC ION | | Authors: | Rajendran, C, Ermler, U, Ludwig, B, Michel, H. | | Deposit date: | 2010-03-20 | | Release date: | 2010-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.332 Å) | | Cite: | Structure at 1.5 A resolution of cytochrome c(552) with its flexible linker segment, a membrane-anchored protein from Paracoccus denitrificans.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
3J2T
 
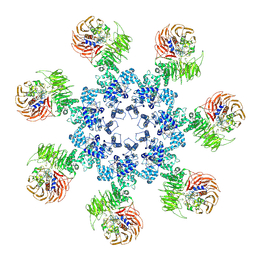 | | An improved model of the human apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Yuan, S, Topf, M, Akey, C.W. | | Deposit date: | 2012-12-23 | | Release date: | 2013-04-10 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Changes in apaf-1 conformation that drive apoptosome assembly.
Biochemistry, 52, 2013
|
|
3HQ9
 
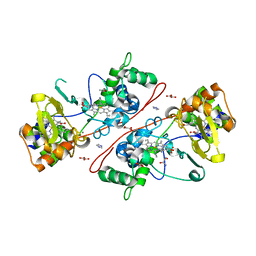 | | CcpA from G. sulfurreducens, S134P variant | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, Cytochrome c551 peroxidase, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ8
 
 | | CcpA from G. sulfurreducens S134P/V135K variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, IMIDAZOLE, ... | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ7
 
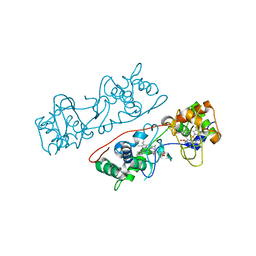 | | CcpA from G. sulfurreducens, G94K/K97Q/R100I variant | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3HQ6
 
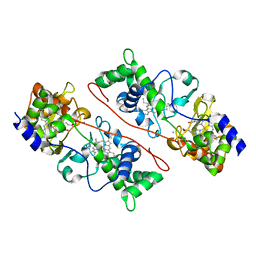 | | Cytochrome c peroxidase from G. sulfurreducens, wild type | | Descriptor: | CALCIUM ION, Cytochrome c551 peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hoffmann, M, Seidel, J, Einsle, O. | | Deposit date: | 2009-06-05 | | Release date: | 2009-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CcpA from Geobacter sulfurreducens is a basic di-heme cytochrome c peroxidase.
J.Mol.Biol., 393, 2009
|
|
3CYT
 
 | |
3CXH
 
 | |
3CX5
 
 | |
3CP5
 
 | | Cytochrome c from rhodothermus marinus | | Descriptor: | Cytochrome c, HEME C, SULFATE ION | | Authors: | Stelter, M, Melo, A, Saraiva, L, Teixeira, M, Archer, M. | | Deposit date: | 2008-03-31 | | Release date: | 2008-10-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | A novel type of monoheme cytochrome c: biochemical and structural characterization at 1.23 A resolution of rhodothermus marinus cytochrome c
Biochemistry, 47, 2008
|
|
3C2C
 
 | |
351C
 
 | |
2ZZS
 
 | | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633 | | Descriptor: | Cytochrome c554, GLYCEROL, HEME C | | Authors: | Akazaki, H, Kawai, F, Kumaki, Y, Sekine, K, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633
To be Published
|
|
2ZXY
 
 | | Crystal Structure of Cytochrome c555 from Aquifex aeolicus | | Descriptor: | Cytochrome c552, HEME C | | Authors: | Obuchi, M, Kawahara, K, Motooka, D, Nakamura, S, Yamanaka, M, Takeda, T, Uchiyama, S, Kobayashi, Y, Ohkubo, T, Sambongi, Y. | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Hyperstability and crystal structure of cytochrome c(555) from hyperthermophilic Aquifex aeolicus
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2ZOO
 
 | | Crystal structure of nitrite reductase from Pseudoalteromonas haloplanktis TAC125 | | Descriptor: | COPPER (II) ION, PROTOPORPHYRIN IX CONTAINING FE, Probable nitrite reductase, ... | | Authors: | Nojiri, M, Tsuda, A, Yamaguchi, K, Suzuki, S. | | Deposit date: | 2008-05-27 | | Release date: | 2009-06-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Electron transfer processes within and between proteins containing the HEME C and blue Cu
To be Published
|
|
2YQB
 
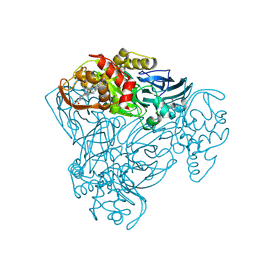 | | Structure of P93A variant of three-domain heme-Cu nitrite reductase from Ralstonia pickettii at 1.4 A resolution | | Descriptor: | COPPER (II) ION, COPPER-CONTAINING NITRITE REDUCTASE, HEME C | | Authors: | Antonyuk, S.V, Han, C, Eady, R.R, Hasnain, S.S. | | Deposit date: | 2012-11-07 | | Release date: | 2013-04-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structures of protein-protein complexes involved in electron transfer.
Nature, 496, 2013
|
|
2YK3
 
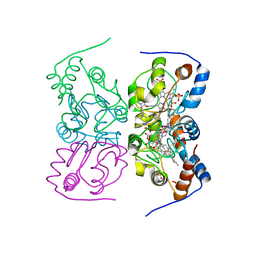 | | CRITHIDIA FASCICULATA CYTOCHROME C | | Descriptor: | CYTOCHROME C, HEME C, SULFATE ION | | Authors: | Fulop, V, Sam, K.A, Ferguson, S.J, Ginger, M.L, Allen, J.W.A. | | Deposit date: | 2011-05-25 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of a Trypanosomatid Mitochondrial Cytochrome C with Heme Attached Via Only One Thioether Bond and Implications for the Substrate Recognition Requirements of Heme Lyase.
FEBS J., 276, 2009
|
|
2YEV
 
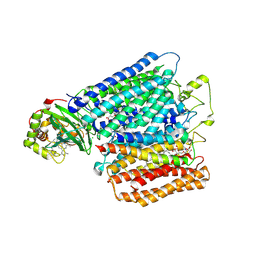 | | Structure of caa3-type cytochrome oxidase | | Descriptor: | (1R,4S,6R)-6-({[2-(ACETYLAMINO)-2-DEOXY-ALPHA-D-GLUCOPYRANOSYL]OXY}METHYL)-4-HYDROXY-1-{[(15-METHYLHEXADECANOYL)OXY]METHYL}-4-OXIDO-7-OXO-3,5-DIOXA-8-AZA-4-PHOSPHAHEPTACOS-1-YL 15-METHYLHEXADECANOATE, (2R)-2,3-DIHYDROXYPROPYL (7Z)-TETRADEC-7-ENOATE, (2R)-3-HYDROXYPROPANE-1,2-DIYL DIHEXADECANOATE, ... | | Authors: | Lyons, J.A, Aragao, D, Soulimane, T, Caffrey, M. | | Deposit date: | 2011-03-31 | | Release date: | 2012-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Structural Insights Into Electron Transfer in Caa3-Type Cytochrome Oxidases.
Nature, 487, 2012
|
|
2YCC
 
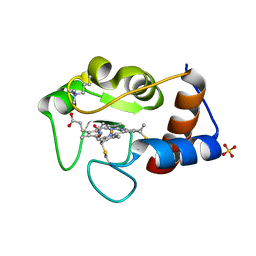 | |
2YBB
 
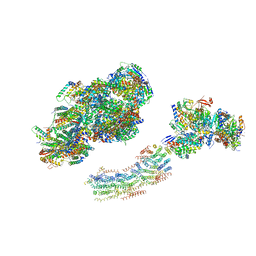 | | Fitted model for bovine mitochondrial supercomplex I1III2IV1 by single particle cryo-EM (EMD-1876) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Althoff, T, Mills, D.J, Popot, J.-L, Kuehlbrandt, W. | | Deposit date: | 2011-03-02 | | Release date: | 2011-10-19 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (19 Å) | | Cite: | Arrangement of Electron Transport Chain Components in Bovine Mitochondrial Supercomplex I(1)III(2)Iv(1).
Embo J., 30, 2011
|
|
2XTS
 
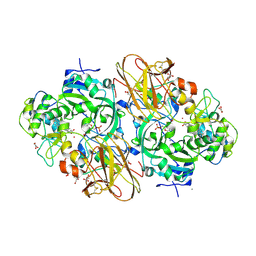 | | Crystal Structure of the Sulfane Dehydrogenase SoxCD from Paracoccus pantotrophus | | Descriptor: | CALCIUM ION, COBALT (II) ION, CYTOCHROME, ... | | Authors: | Zander, U, Faust, A, Klink, B.U, de Sanctis, D, Panjikar, S, Quentmeier, A, Bardischewski, F, Friedrich, C.G, Scheidig, A.J. | | Deposit date: | 2010-10-12 | | Release date: | 2010-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structural Basis for the Oxidation of Protein-Bound Sulfur by the Sulfur Cycle Molybdohemo-Enzyme Sulfane Dehydrogenase Soxcd.
J.Biol.Chem., 286, 2011
|
|
