5HRN
 
 | | HIV Integrase Catalytic Domain containing F185K mutation complexed with GSK0002 | | Descriptor: | (2S)-tert-butoxy[1-(3,4-difluorobenzyl)-6-methyl-4-(5-methyl-3,4-dihydro-2H-chromen-6-yl)-1H-pyrrolo[2,3-b]pyridin-5-yl]acetic acid, 1,2-ETHANEDIOL, CACODYLATE ION, ... | | Authors: | Nolte, R.T. | | Deposit date: | 2016-01-23 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Inhibitor-Induced Aggregation of HIV Integrase.
PLoS Biol., 14, 2016
|
|
5TEG
 
 | |
5H0O
 
 | | Crystal structure of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease with manganese ion | | Descriptor: | HNH endonuclease, MANGANESE (II) ION | | Authors: | Zhang, L.K, Xu, D.D, Huang, Y.C, Gong, Y. | | Deposit date: | 2016-10-06 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Structural and functional characterization of deep-sea thermophilic bacteriophage GVE2 HNH endonuclease.
Sci Rep, 7, 2017
|
|
1SV5
 
 | | CRYSTAL STRUCTURE OF K103N MUTANT HIV-1 REVERSE TRANSCRIPTASE (RT) IN COMPLEX WITH JANSSEN-R165335 | | Descriptor: | 4-({6-AMINO-5-BROMO-2-[(4-CYANOPHENYL)AMINO]PYRIMIDIN-4-YL}OXY)-3,5-DIMETHYLBENZONITRILE, Reverse Transcriptase | | Authors: | Das, K, Arnold, E. | | Deposit date: | 2004-03-27 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Roles of Conformational and Positional Adaptability in Structure-Based Design of TMC125-R165335 (Etravirine) and Related Non-nucleoside Reverse Transcriptase Inhibitors That Are Highly Potent and Effective against Wild-Type and Drug-Resistant HIV-1 Variants
J.Med.Chem., 47, 2004
|
|
2B5L
 
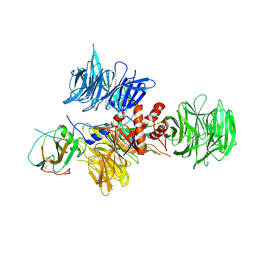 | | Crystal Structure of DDB1 In Complex with Simian Virus 5 V Protein | | Descriptor: | Nonstructural protein V, ZINC ION, damage-specific DNA binding protein 1 | | Authors: | Li, T, Chen, X, Garbutt, K.C, Zhou, P, Zheng, N. | | Deposit date: | 2005-09-28 | | Release date: | 2006-02-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase.
Cell(Cambridge,Mass.), 124, 2006
|
|
1CJT
 
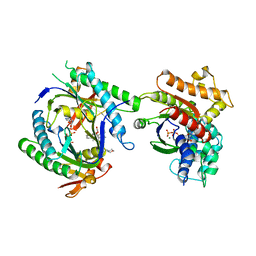 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH BETA-L-2',3'-DIDEOXYATP, MN, AND MG | | Descriptor: | 2',3'-DIDEOXYADENOSINE-5'-TRIPHOSPHATE, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
4P8O
 
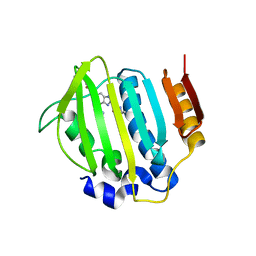 | | S. aureus gyrase bound to an aminobenzimidazole urea inhibitor | | Descriptor: | 1-ethyl-3-[5-(5-fluoropyridin-3-yl)-7-(pyrimidin-2-yl)-1H-benzimidazol-2-yl]urea, DNA gyrase subunit B | | Authors: | Jacobs, M.D. | | Deposit date: | 2014-03-31 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Second-generation antibacterial benzimidazole ureas: discovery of a preclinical candidate with reduced metabolic liability.
J.Med.Chem., 57, 2014
|
|
1CJK
 
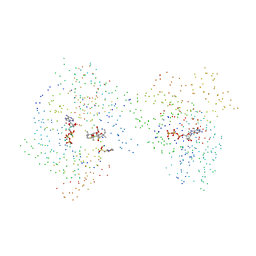 | | COMPLEX OF GS-ALPHA WITH THE CATALYTIC DOMAINS OF MAMMALIAN ADENYLYL CYCLASE: COMPLEX WITH ADENOSINE 5'-(ALPHA THIO)-TRIPHOSPHATE (RP), MG, AND MN | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, ADENOSINE-5'-RP-ALPHA-THIO-TRIPHOSPHATE, ... | | Authors: | Tesmer, J.J.G, Sprang, S.R. | | Deposit date: | 1999-04-16 | | Release date: | 1999-08-31 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Two-metal-Ion catalysis in adenylyl cyclase.
Science, 285, 1999
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | Descriptor: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | Authors: | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | Deposit date: | 1998-06-30 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
4PWM
 
 | | Crystal structure of Dickerson Drew Dodecamer with 5-carboxycytosine | | Descriptor: | 5'-[CGCGAATT(5CC)GCG]-3' | | Authors: | Szulik, M.W, Pallan, P, Banerjee, S, Voehler, M, Egli, M, Stone, M.P. | | Deposit date: | 2014-03-20 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Differential stabilities and sequence-dependent base pair opening dynamics of watson-crick base pairs with 5-hydroxymethylcytosine, 5-formylcytosine, or 5-carboxylcytosine.
Biochemistry, 54, 2015
|
|
3VOW
 
 | | Crystal Structure of the Human APOBEC3C having HIV-1 Vif-binding Interface | | Descriptor: | CHLORIDE ION, Probable DNA dC->dU-editing enzyme APOBEC-3C, ZINC ION | | Authors: | Kitamura, S, Suzuki, A, Watanabe, N, Iwatani, Y. | | Deposit date: | 2012-02-22 | | Release date: | 2012-10-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The APOBEC3C crystal structure and the interface for HIV-1 Vif binding.
Nat.Struct.Mol.Biol., 19, 2012
|
|
3QLH
 
 | | HIV-1 Reverse Transcriptase in Complex with Manicol at the RNase H Active Site and TMC278 (Rilpivirine) at the NNRTI Binding Pocket | | Descriptor: | (2S)-5,7-dihydroxy-9-methyl-2-(prop-1-en-2-yl)-1,2,3,4-tetrahydro-6H-benzo[7]annulen-6-one, 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, ... | | Authors: | Himmel, D.M, Wojtak, K, Bauman, J.D, Arnold, E. | | Deposit date: | 2011-02-02 | | Release date: | 2011-12-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H.
J.Med.Chem., 54, 2011
|
|
1KZ2
 
 | | Solution structure of the third helix of Antennapedia homeodomain derivative [W6F,W14F] | | Descriptor: | Antennapedia protein | | Authors: | Czajlik, A, Mesko, E, Penke, B, Perczel, A. | | Deposit date: | 2002-02-06 | | Release date: | 2002-02-20 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Investigation of penetratin peptides. Part 1. The environment dependent conformational properties of penetratin and two of its derivatives.
J.Pept.Sci., 8, 2002
|
|
1KZ0
 
 | |
1H3O
 
 | | Crystal Structure of the Human TAF4-TAF12 (TAFII135-TAFII20) Complex | | Descriptor: | TRANSCRIPTION INITIATION FACTOR TFIID 135 KDA SUBUNIT, TRANSCRIPTION INITIATION FACTOR TFIID 20/15 KDA SUBUNITS | | Authors: | Werten, S, Mitschler, A, Moras, D. | | Deposit date: | 2002-09-12 | | Release date: | 2002-09-26 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Subcomplex of Human Transcription Factor TFIID Formed by TATA Binding Protein-Associated Factors Htaf4 (Htaf(II)135) and Htaf12 (Htaf(II)20).
J.Biol.Chem., 277, 2002
|
|
2FCO
 
 | | Crystal Structure of Bacillus stearothermophilus PrfA-Holliday Junction Resolvase | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, recombination protein U (penicillin-binding protein related factor A) | | Authors: | Li, J, Jedrzejas, M.J. | | Deposit date: | 2005-12-12 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure, flexibility, and mechanism of the Bacillus stearothermophilus RecU Holliday junction resolvase.
Proteins, 68, 2007
|
|
5XBL
 
 | | Structure of nuclease in complex with associated protein | | Descriptor: | Associated protein, CRISPR-associated endonuclease Cas9/Csn1, RNA (98-MER) | | Authors: | Dong, D, Guo, M, Wang, S, Zhu, Y, Huang, Z. | | Deposit date: | 2017-03-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.052 Å) | | Cite: | Structural basis of CRISPR-SpyCas9 inhibition by an anti-CRISPR protein
Nature, 546, 2017
|
|
7MSV
 
 | |
8QEK
 
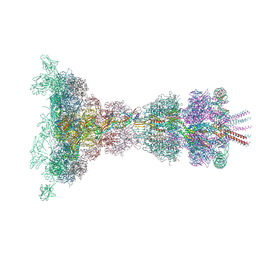 | | Neck and tail of phage 812 after tail contraction (composite) | | Descriptor: | Anchor DNA forward strand (120-MER), Anchor DNA reverse strand (120-MER), Baseplate hub assembly protein, ... | | Authors: | Cienikova, Z, Siborova, M, Fuzik, T, Plevka, P. | | Deposit date: | 2023-08-31 | | Release date: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Genome anchoring, retention, and release by neck proteins of Herelleviridae phage 812
To Be Published
|
|
7MPZ
 
 | |
6Y2D
 
 | | Crystal structure of the second KH domain of FUBP1 | | Descriptor: | Far upstream element-binding protein 1, GLYCEROL, SULFATE ION | | Authors: | Ni, X, Chaikuad, A, Joerger, A.C, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
6Y2C
 
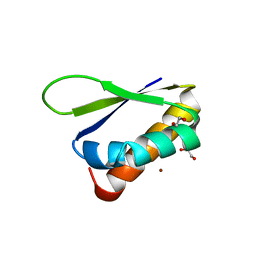 | | Crystal structure of the third KH domain of FUBP1 | | Descriptor: | 1,2-ETHANEDIOL, Far upstream element-binding protein 1, ZINC ION | | Authors: | Ni, X, Joerger, A.C, Chaikuad, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-02-15 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Comparative structural analyses and nucleotide-binding characterization of the four KH domains of FUBP1.
Sci Rep, 10, 2020
|
|
7AJT
 
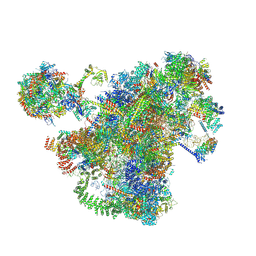 | | Cryo-EM structure of the 90S-exosome super-complex (state Pre-A1-exosome) | | Descriptor: | 13 kDa ribonucleoprotein-associated protein, 18S rRNA, 40S ribosomal protein S1-A, ... | | Authors: | Cheng, J, Lau, B, Flemming, D, Venuta, G.L, Berninghausen, O, Beckmann, R, Hurt, E. | | Deposit date: | 2020-09-29 | | Release date: | 2020-12-30 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the Maturing 90S Pre-ribosome in Association with the RNA Exosome.
Mol.Cell, 81, 2021
|
|
8BR9
 
 | |
8RKU
 
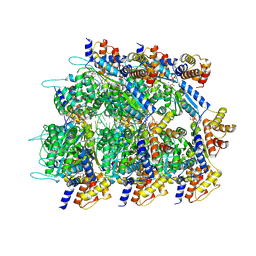 | | Conformational Landscape of the Type V-K CRISPR-associated TransposonIntegration Assembly CAST V-K TnsC domain local-refinement map | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Non-target strand - LE, ... | | Authors: | Tenjo-Castano, F, Mesa, P, Montoya, G. | | Deposit date: | 2023-12-30 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Conformational landscape of the type V-K CRISPR-associated transposon integration assembly.
Mol.Cell, 84, 2024
|
|
