6JCR
 
 | | AAV1 in neutral condition at 3.07 Ang | | Descriptor: | Capsid protein | | Authors: | Lou, Z, Zhang, R. | | Deposit date: | 2019-01-30 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Divergent engagements between adeno-associated viruses with their cellular receptor AAVR.
Nat Commun, 10, 2019
|
|
6SD4
 
 | | Structure of the RBM3/collar region of the Salmonella flagella MS-ring protein FliF with 34-fold symmetry applied | | Descriptor: | Flagellar M-ring protein | | Authors: | Johnson, S, Fong, Y.H, Deme, J.C, Furlong, E.J, Kuhlen, L, Lea, S.M. | | Deposit date: | 2019-07-26 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Symmetry mismatch in the MS-ring of the bacterial flagellar rotor explains the structural coordination of secretion and rotation.
Nat Microbiol, 5, 2020
|
|
1WZI
 
 | |
7C1Z
 
 | | ATP bound structure of Pseudouridine kinase (PUKI) from Arabidopsis thaliana | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PfkB-like carbohydrate kinase family protein, ... | | Authors: | Kim, S.H, Rhee, S. | | Deposit date: | 2020-05-06 | | Release date: | 2020-11-18 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.09617043 Å) | | Cite: | Structural basis for the substrate specificity and catalytic features of pseudouridine kinase from Arabidopsis thaliana.
Nucleic Acids Res., 49, 2021
|
|
8R3C
 
 | |
6B6B
 
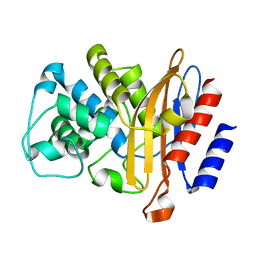 | |
8G6I
 
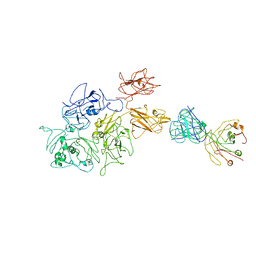 | | Coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Coagulation factor VIII chimera from human and pig, NB33 heavy chain, ... | | Authors: | Childers, K.C, Davulcu, O, Haynes, R.M, Lollar, P, Doering, C.B, Coxon, C.H, Spiegel, P.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure of coagulation factor VIII bound to a patient-derived anti-C1 domain antibody inhibitor.
Blood, 142, 2023
|
|
1TTI
 
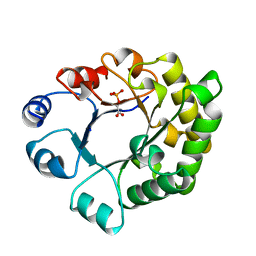 | |
6MS4
 
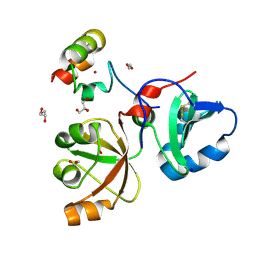 | | Crystal structure of the DENR-MCT-1 complex | | Descriptor: | Density-regulated protein, GLYCEROL, Malignant T-cell-amplified sequence 1, ... | | Authors: | Lomakin, I.B, Steitz, T.A, Dmitriev, S.E. | | Deposit date: | 2018-10-16 | | Release date: | 2019-01-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Crystal structure of the DENR-MCT-1 complex revealed zinc-binding site essential for heterodimer formation.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
7BTL
 
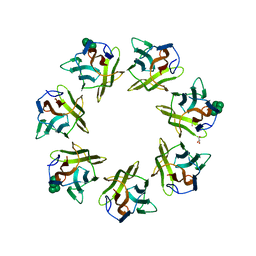 | | Mevo lectin complex with mannopentose | | Descriptor: | GLYCEROL, alpha-D-mannopyranose, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose, ... | | Authors: | Sivaji, N, Suguna, K, Surolia, A, Vijayan, M. | | Deposit date: | 2020-04-01 | | Release date: | 2021-02-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and related studies on Mevo lectin from Methanococcus voltae A3: the first thorough characterization of an archeal lectin and its interactions.
Glycobiology, 31, 2021
|
|
6J9B
 
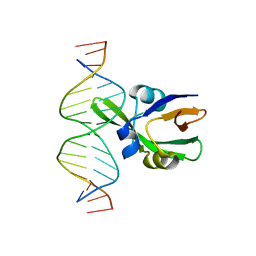 | | Arabidopsis FUS3-DNA complex | | Descriptor: | B3 domain-containing transcription factor FUS3, DNA (5'-D(*AP*AP*TP*CP*CP*AP*TP*GP*CP*AP*GP*AP*AP*TP*C)-3'), DNA (5'-D(*AP*TP*TP*CP*TP*GP*CP*AP*TP*GP*GP*AP*TP*TP*G)-3') | | Authors: | Hu, H, Du, J. | | Deposit date: | 2019-01-22 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Embryonic resetting of the parental vernalized state by two B3 domain transcription factors in Arabidopsis.
Nat.Plants, 5, 2019
|
|
7BWX
 
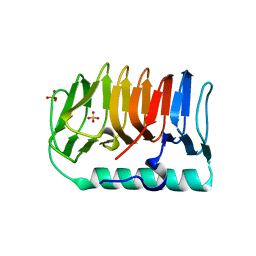 | | Crystal structure of ice-binding protein from an Antarctic ascomycete, Antarctomyces psychrotrophicus. | | Descriptor: | GLYCEROL, Ice-binding protein isoform1a, SULFATE ION | | Authors: | Yamauchi, A, Arai, T, Kondo, H, Tsuda, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.904 Å) | | Cite: | An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes.
Biomolecules, 10, 2020
|
|
8SBH
 
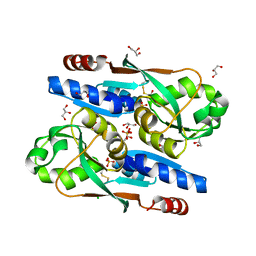 | | YeiE effector binding domain from E. coli | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Momany, C, Nune, M, Brondani, J.C, Afful, D, Neidle, E. | | Deposit date: | 2023-04-03 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | FinR, a LysR-type transcriptional regulator involved in sulfur homeostasis with homologs in diverse microorganisms
To Be Published
|
|
6B9F
 
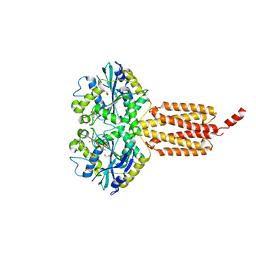 | | Human ATL1 mutant - F151S bound to GDPAlF4- | | Descriptor: | Atlastin-1, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | O'Donnell, J.P, Sondermann, H. | | Deposit date: | 2017-10-10 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A hereditary spastic paraplegia-associated atlastin variant exhibits defective allosteric coupling in the catalytic core.
J. Biol. Chem., 293, 2018
|
|
7L3L
 
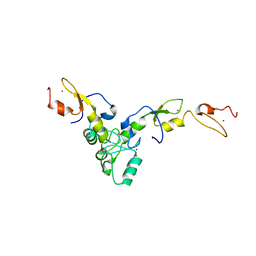 | | Structure of TRAF5 and TRAF6 RING Hetero dimer | | Descriptor: | TNF receptor-associated factor 5, TNF receptor-associated factor 6, ZINC ION | | Authors: | Das, A, Middleton, A.J, Padala, P, Day, C.L. | | Deposit date: | 2020-12-17 | | Release date: | 2021-02-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure and ubiquitin binding properties of TRAF RING heterodimers.
J.Mol.Biol., 2021
|
|
6JC0
 
 | |
8G5D
 
 | |
5MEC
 
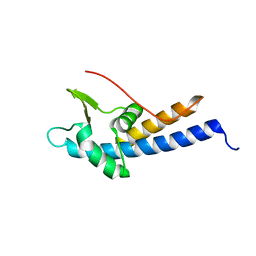 | |
6SW5
 
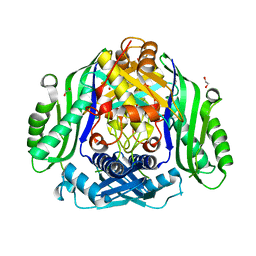 | | Crystal structure of the human S-adenosylmethionine synthetase 1 (ligand-free form) | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, S-adenosylmethionine synthase isoform type-1 | | Authors: | Panmanee, J, Antoyuk, S.V, Hasnain, S.S. | | Deposit date: | 2019-09-19 | | Release date: | 2020-06-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of the dominant inheritance of hypermethioninemia associated with the Arg264His mutation in the MAT1A gene.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
5JGP
 
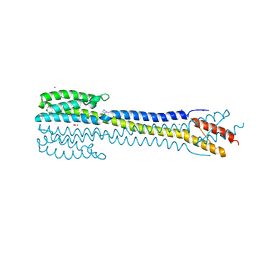 | | Crystal structure of the nitrate/nitrite sensor NarQ fragment bound with iodide ions | | Descriptor: | IODIDE ION, NITRATE ION, Nitrate/nitrite sensor protein NarQ | | Authors: | Melnikov, I, Polovinkin, V, Popov, A, Gordeliy, V. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-31 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Fast iodide-SAD phasing for high-throughput membrane protein structure determination.
Sci Adv, 3, 2017
|
|
7BWY
 
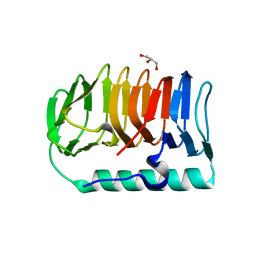 | | Crystal structure of ice-binding protein from an Antarctic ascomycete, Antarctomyces psychrotrophicus. | | Descriptor: | GLYCEROL, Ice-binding protein isoform1a | | Authors: | Yamauchi, A, Arai, T, Kondo, H, Tsuda, S. | | Deposit date: | 2020-04-16 | | Release date: | 2020-05-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | An Ice-Binding Protein from an Antarctic Ascomycete Is Fine-Tuned to Bind to Specific Water Molecules Located in the Ice Prism Planes.
Biomolecules, 10, 2020
|
|
5MF7
 
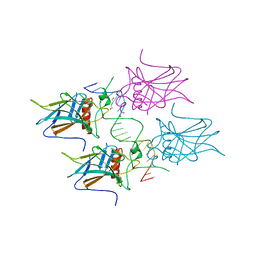 | | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins (complex p53DBD-GADD45) | | Descriptor: | Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, DNA, ... | | Authors: | Rozenberg, H, Diskin-Posner, Y, Golovenko, D, Shakked, Z. | | Deposit date: | 2016-11-17 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | New Insights into the Role of DNA Shape on Its Recognition by p53 Proteins.
Structure, 26, 2018
|
|
1BMD
 
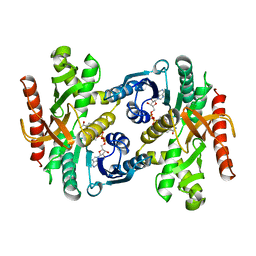 | |
7KMZ
 
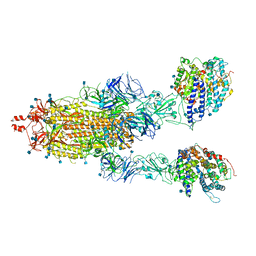 | | Cryo-EM structure of double ACE2-bound SARS-CoV-2 trimer Spike at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Gorman, J, Kwong, P.D, Shapiro, L. | | Deposit date: | 2020-11-03 | | Release date: | 2020-12-09 | | Last modified: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains.
Cell Host Microbe, 28, 2020
|
|
8AXX
 
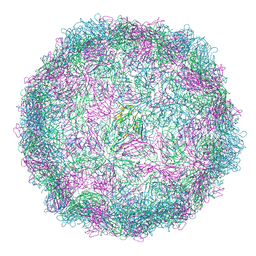 | | Expanded Coxsackievirus A9 after treatment with endosomal ionic buffer | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3 | | Authors: | Domanska, A, Plavec, Z, Ruokolainen, V, Loflund, B, Marjomaki, V.S, Butcher, S.J. | | Deposit date: | 2022-09-01 | | Release date: | 2022-10-19 | | Last modified: | 2023-02-01 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural Studies Reveal that Endosomal Cations Promote Formation of Infectious Coxsackievirus A9 A-Particles, Facilitating RNA and VP4 Release.
J.Virol., 96, 2022
|
|
