6NOM
 
 | | NMR solution structure of Pisum sativum defensin 2 (Psd2) provides evidence for the presence of hydrophobic surface clusters | | Descriptor: | Defensin-2 | | Authors: | Pinheiro-Aguiar, R, Amaral, V.S.G, Bastos, I, Kurtenbach, E, Almeida, F.C.L. | | Deposit date: | 2019-01-16 | | Release date: | 2019-08-21 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of Pisum sativum defensin 2 provides evidence for the presence of hydrophobic surface-clusters.
Proteins, 88, 2020
|
|
6PV2
 
 | |
6PV0
 
 | |
1BJE
 
 | |
6WW7
 
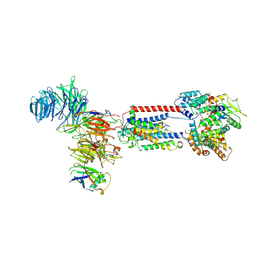 | | Structure of the human ER membrane protein complex in a lipid nanodisc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ER Membrane Protein Complex Subunit 4, ... | | Authors: | Tomaleri, G.P, Januszyk, K, Pleiner, T, Inglis, A.J, Voorhees, R.M. | | Deposit date: | 2020-05-08 | | Release date: | 2020-05-27 | | Last modified: | 2020-08-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for membrane insertion by the human ER membrane protein complex.
Science, 369, 2020
|
|
2HSL
 
 | | NMR structure of 13mer duplex DNA containing an abasic site, averaged structure (alpha anomer) | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(D1P)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
2HSS
 
 | | 13mer duplex DNA containg an abasic site with beta anomer, averaged structure | | Descriptor: | 5'-D(*CP*CP*AP*AP*AP*GP*(AAB)P*AP*CP*CP*GP*GP*G)-3', 5'-D(*CP*CP*CP*GP*GP*TP*AP*CP*TP*TP*TP*GP*G)-3' | | Authors: | Chen, J, Dupradeau, F.Y, Case, D.A, Turner, C.J, Stubbe, J. | | Deposit date: | 2006-07-22 | | Release date: | 2007-05-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance structural studies and molecular modeling of duplex DNA containing normal and 4'-oxidized abasic sites.
Biochemistry, 46, 2007
|
|
1BOF
 
 | | GI-ALPHA-1 BOUND TO GDP AND MAGNESIUM | | Descriptor: | GI ALPHA 1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Coleman, D.E, Sprang, S.R. | | Deposit date: | 1998-08-04 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of the G protein Gi alpha 1 complexed with GDP and Mg2+: a crystallographic titration experiment.
Biochemistry, 37, 1998
|
|
6UD0
 
 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
6QYT
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - Mutacin I Ring A truncated analogue | | Descriptor: | DAL-LEU-SER-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
1D4L
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | (10S,13S,1'R)-13-[1'-HYDROXY-2'-(N-P-AMINOBENZENESULFONYL-1''-AMINO-3''-METHYLBUTYL)ETHYL]-8,11-DIOXO-10-ISOPROPYL-2-OXA-9,12-DIAZABICYCLO [13.2.2]NONADECA-15,17,18-TRIENE, HIV-1 PROTEASE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
6U46
 
 | |
1D8F
 
 | | CRYSTAL STRUCTURE OF MMP3 COMPLEXED WITH A PIPERAZINE BASED INHIBITOR. | | Descriptor: | CALCIUM ION, N-HYDROXY-1-(4-METHOXYPHENYL)SULFONYL-4-BENZYLOXYCARBONYL-PIPERAZINE-2-CARBOXAMIDE, STROMELYSIN-1 PRECURSOR, ... | | Authors: | Cheng, M.Y, De, B, Pikul, S, Almstead, N.G, Natchus, M.G, Anastasio, M.V, McPhail, S.J, Snider, C.E, Taiwo, Y.O, Chen, L.Y. | | Deposit date: | 1999-10-22 | | Release date: | 2000-10-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and synthesis of piperazine-based matrix metalloproteinase inhibitors.
J.Med.Chem., 43, 2000
|
|
6UCO
 
 | |
8RJX
 
 | |
6UCP
 
 | |
8QTQ
 
 | | Thermostable WW domain | | Descriptor: | WW domain | | Authors: | Kovermann, M, Thomas, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-07-03 | | Method: | SOLUTION NMR | | Cite: | Thermostable WW-Domain Scaffold to Design Functional beta-Sheet Miniproteins.
J.Am.Chem.Soc., 2024
|
|
1D4K
 
 | | HIV-1 PROTEASE COMPLEXED WITH A MACROCYCLIC PEPTIDOMIMETIC INHIBITOR | | Descriptor: | HIV-1 PROTEASE, N-13-[(10S,13S)-9,12-DIOXO-10-(2-BUTYL)-2-OXA-8,11-DIAZABICYCLO [13.2.2] NONADECA-15,17,18-TRIENE] (2R)-BENZYL-(4S)-HYDROXY-5-AMINOPENTANOIC (1R)-HYDROXY-(2S)-INDANEAMIDE, SULFATE ION | | Authors: | Tyndall, J.D, Reid, R.C, Tyssen, D.P, Jardine, D.K, Todd, B, Passmore, M, March, D.R, Pattenden, L.K, Alewood, D, Hu, S.H, Alewood, P.F, Birch, C.J, Martin, J.L, Fairlie, D.P. | | Deposit date: | 1999-10-04 | | Release date: | 2000-10-11 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis, stability, antiviral activity, and protease-bound structures of substrate-mimicking constrained macrocyclic inhibitors of HIV-1 protease.
J.Med.Chem., 43, 2000
|
|
8K20
 
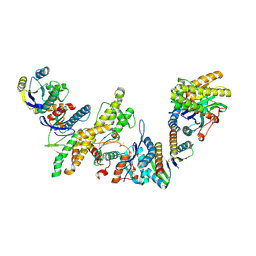 | | Cryo-EM structure of KEOPS complex from Arabidopsis thaliana | | Descriptor: | At4g34412, At5g53043, FE (III) ION, ... | | Authors: | Zheng, X.X, Zhu, L, Duan, L, Zhang, W.H. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A.
Nucleic Acids Res., 52, 2024
|
|
6WXE
 
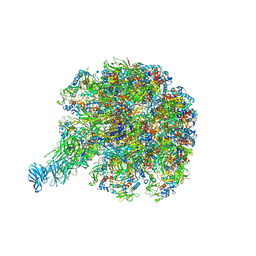 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Upright conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6QYW
 
 | | Solution NMR of synthetic analogues of nisin and mutacin ring A and ring B - nisin ring A | | Descriptor: | ILE-DBU-DAL-ILE-DHA-LEU-CYS-ALA | | Authors: | Dickman, R, Mitchell, S.A, Figueiredo, A, Hansen, D.F, Tabor, A.B. | | Deposit date: | 2019-03-09 | | Release date: | 2019-09-11 | | Last modified: | 2019-10-02 | | Method: | SOLUTION NMR | | Cite: | Molecular Recognition of Lipid II by Lantibiotics: Synthesis and Conformational Studies of Analogues of Nisin and Mutacin Rings A and B.
J.Org.Chem., 84, 2019
|
|
7UD0
 
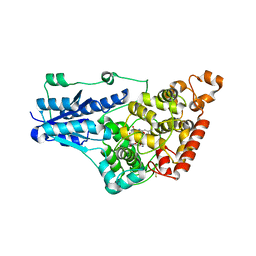 | | Room Temperature Drosophila Cryptochrome | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Schneps, C.M, Crane, B.R. | | Deposit date: | 2022-03-17 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Room-temperature serial synchrotron crystallography of Drosophila cryptochrome.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6VSA
 
 | |
7UBI
 
 | |
1B3P
 
 | | 5'-D(*GP*GP*AP*GP*GP*AP*T)-3' | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*GP*AP*T)-3') | | Authors: | Kettani, A, Bouaziz, S, Skripkin, E, Majumdar, A, Wang, W, Jones, R.A, Patel, D.J. | | Deposit date: | 1998-12-14 | | Release date: | 1999-08-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Interlocked mismatch-aligned arrowhead DNA motifs.
Structure Fold.Des., 7, 1999
|
|
