5TJH
 
 | | hUGDH A136M Substitution | | Descriptor: | CHLORIDE ION, PYROPHOSPHATE 2-, UDP-glucose 6-dehydrogenase, ... | | Authors: | Beattie, N.R, Wood, Z.A. | | Deposit date: | 2016-10-04 | | Release date: | 2016-11-02 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allostery and Hysteresis Are Coupled in Human UDP-Glucose Dehydrogenase.
Biochemistry, 56, 2017
|
|
5TE1
 
 | |
5TFZ
 
 | |
5T73
 
 | |
5TCY
 
 | | A complex of the synthetic siderophore analogue Fe(III)-5-LICAM with CeuE (H227L variant), a periplasmic protein from Campylobacter jejuni. | | Descriptor: | Enterochelin uptake periplasmic binding protein, FE (III) ION, N,N'-pentane-1,5-diylbis(2,3-dihydroxybenzamide) | | Authors: | Wilde, E.J, Blagova, E, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-09-16 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
5TEK
 
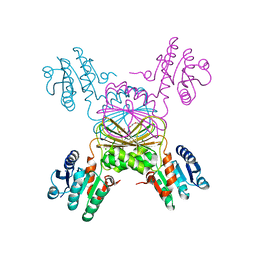 | | Apo Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Mycobacterium tuberculosis | | Descriptor: | 4-hydroxy-tetrahydrodipicolinate reductase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Mank, N, Pote, S, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-21 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
5SY6
 
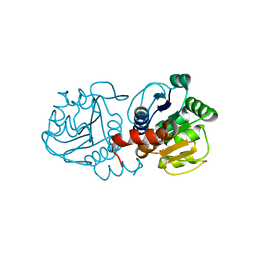 | | Atomic resolution structure of human DJ-1, DTT bound | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, Protein deglycase DJ-1 | | Authors: | Wilson, M.A, Lin, J. | | Deposit date: | 2016-08-10 | | Release date: | 2016-12-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Short Carboxylic Acid-Carboxylate Hydrogen Bonds Can Have Fully Localized Protons.
Biochemistry, 56, 2017
|
|
5T1A
 
 | | Structure of CC Chemokine Receptor 2 with Orthosteric and Allosteric Antagonists | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2~{R})-1-(4-chloranyl-2-fluoranyl-phenyl)-2-cyclohexyl-3-ethanoyl-4-oxidanyl-2~{H}-pyrrol-5-one, (3S)-1-{(1S,2R,4R)-4-[methyl(propan-2-yl)amino]-2-propylcyclohexyl}-3-{[6-(trifluoromethyl)quinazolin-4-yl]amino}pyrrolidin-2-one, ... | | Authors: | Zheng, Y, Qin, L, Ortiz Zacarias, N.V, de Vries, H, Han, G.W, Gustavsson, M, Dabros, M, Zhao, C, Cherney, R.J, Carter, P, Stamos, D, Abagyan, R, Cherezov, V, Stevens, R.C, IJzerman, A.P, Heitman, L.H, Tebben, A, Kufareva, I, Handel, T.M. | | Deposit date: | 2016-08-18 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.806 Å) | | Cite: | Structure of CC chemokine receptor 2 with orthosteric and allosteric antagonists.
Nature, 540, 2016
|
|
7CB9
 
 | |
5T31
 
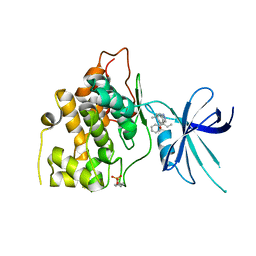 | | Exploiting an Asp-Glu switch in Glycogen Synthase Kinase 3 to design paralog selective inhibitors for use in acute myeloid leukemia | | Descriptor: | (4~{S})-4-ethyl-7,7-dimethyl-4-phenyl-2,6,8,9-tetrahydropyrazolo[3,4-b]quinolin-5-one, Glycogen synthase kinase-3 beta | | Authors: | Stein, A.J, Holson, E.B, Wagner, F.F, Cambell, A.J. | | Deposit date: | 2016-08-24 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Exploiting an Asp-Glu "switch" in glycogen synthase kinase 3 to design paralog-selective inhibitors for use in acute myeloid leukemia.
Sci Transl Med, 10, 2018
|
|
5T5I
 
 | | TUNGSTEN-CONTAINING FORMYLMETHANOFURAN DEHYDROGENASE FROM METHANOTHERMOBACTER WOLFEII, ORTHORHOMBIC FORM AT 1.9 A | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, CALCIUM ION, GLYCEROL, ... | | Authors: | Wagner, T, Ermler, U, Shima, S. | | Deposit date: | 2016-08-31 | | Release date: | 2016-10-19 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The methanogenic CO2 reducing-and-fixing enzyme is bifunctional and contains 46 [4Fe-4S] clusters.
Science, 354, 2016
|
|
5T7N
 
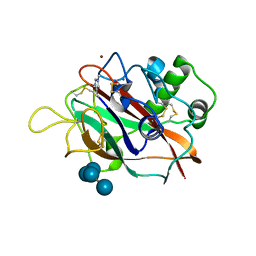 | | X-ray crystal structure of AA13 LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, ZINC ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Tovborg, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2016-09-05 | | Release date: | 2017-01-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Learning from oligosaccharide soaks of crystals of an AA13 lytic polysaccharide monooxygenase: crystal packing, ligand binding and active-site disorder.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7UQ0
 
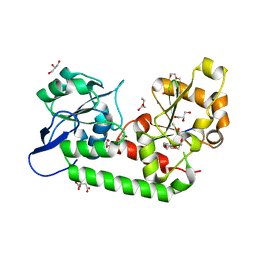 | | Putative periplasmic iron siderophore binding protein FecB (Rv3044) from Mycobacterium tuberculosis | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Chao, A, Cuthbert, B.J, Goulding, C.W. | | Deposit date: | 2022-04-18 | | Release date: | 2022-10-05 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differentiating the roles of Mycobacterium tuberculosis substrate binding proteins, FecB and FecB2, in iron uptake.
Plos Pathog., 19, 2023
|
|
6DA8
 
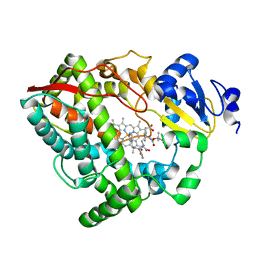 | | Human CYP3A4 bound to an inhibitor | | Descriptor: | Cytochrome P450 3A4, Nalpha-{(2S)-2-[(tert-butoxycarbonyl)amino]-3-phenylpropyl}-N-[(pyridin-3-yl)methyl]-D-phenylalaninamide, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Sevrioukova, I.F. | | Deposit date: | 2018-05-01 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.802 Å) | | Cite: | Structure-Activity Relationships of Rationally Designed Ritonavir Analogues: Impact of Side-Group Stereochemistry, Headgroup Spacing, and Backbone Composition on the Interaction with CYP3A4.
Biochemistry, 58, 2019
|
|
3AOA
 
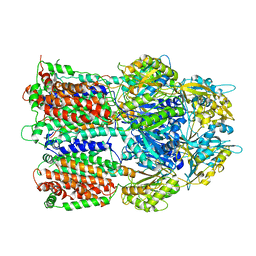 | |
5C3M
 
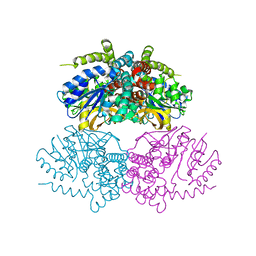 | | Crystal structure of Gan4C, a GH4 6-phospho-glucosidase from Geobacillus stearothermophilus | | Descriptor: | MANGANESE (II) ION, Putative 6-phospho-beta-glucosidase | | Authors: | Cohen, T, Lansky, S, Zehavi, A, Shoham, Y, Shoham, G. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.059 Å) | | Cite: | Crystal structure of Gan4C, a GH4 6-phospho-glucosidase from Geobacillus stearothermophilus
To Be Published
|
|
5SUN
 
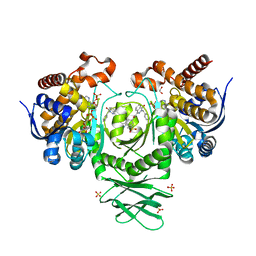 | | IDH1 R132H in complex with IDH146 | | Descriptor: | 1,2-ETHANEDIOL, 3-benzyl-N-[3-(dimethylsulfamoyl)phenyl]-4-oxo-3,4-dihydrophthalazine-1-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Xie, X, Kulathila, R. | | Deposit date: | 2016-08-03 | | Release date: | 2017-02-08 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Allosteric Mutant IDH1 Inhibitors Reveal Mechanisms for IDH1 Mutant and Isoform Selectivity.
Structure, 25, 2017
|
|
4BWB
 
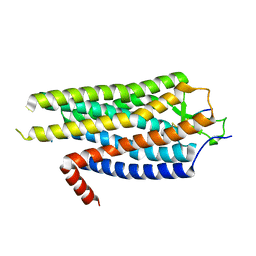 | | Structure of Evolved Agonist-bound Neurotensin Receptor 1 Mutant without Lysozyme Fusion | | Descriptor: | NEUROTENSIN, NEUROTENSIN RECEPTOR TYPE 1 | | Authors: | Egloff, P, Hillenbrand, M, Scott, D.J, Schlinkmann, K.M, Heine, P, Balada, S, Batyuk, A, Mittl, P, Plueckthun, A. | | Deposit date: | 2013-07-01 | | Release date: | 2014-01-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.57 Å) | | Cite: | Structure of Signaling-Competent Neurotensin Receptor 1 Obtained by Directed Evolution in Escherichia Coli
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5C4I
 
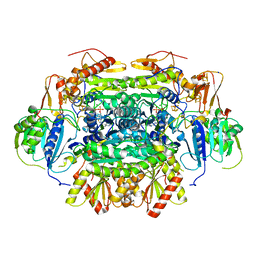 | | Structure of an Oxalate Oxidoreductase | | Descriptor: | IRON/SULFUR CLUSTER, MAGNESIUM ION, Oxalate oxidoreductase subunit alpha, ... | | Authors: | Gibson, M.I, Brignole, E.J, Pierce, E, Can, M, Ragsdale, S.W, Drennan, C.L. | | Deposit date: | 2015-06-18 | | Release date: | 2015-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.274 Å) | | Cite: | The Structure of an Oxalate Oxidoreductase Provides Insight into Microbial 2-Oxoacid Metabolism.
Biochemistry, 54, 2015
|
|
5T99
 
 | |
5T9X
 
 | | Crystal structure of BuGH16Bwt | | Descriptor: | Glycoside Hydrolase, IMIDAZOLE, SODIUM ION | | Authors: | Pluvinage, B, Boraston, A.B. | | Deposit date: | 2016-09-09 | | Release date: | 2017-09-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular basis of an agarose metabolic pathway acquired by a human intestinal symbiont.
Nat Commun, 9, 2018
|
|
5TDE
 
 | | TEV Cleaved Human ATP Citrate Lyase Bound to Citrate | | Descriptor: | ATP-citrate synthase, CITRATE ANION, DIHYDROGENPHOSPHATE ION, ... | | Authors: | Hu, J, Fraser, M.E. | | Deposit date: | 2016-09-19 | | Release date: | 2017-08-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding of hydroxycitrate to human ATP-citrate lyase.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5TDV
 
 | | Intermediate O2 diiron complex in the Q228A variant of Toluene 4-moonoxygenase (T4moHD) | | Descriptor: | FE (III) ION, PEROXIDE ION, Toluene-4-monooxygenase system protein A, ... | | Authors: | Bailey, L.J, Acheson, J.F, Fox, B.G. | | Deposit date: | 2016-09-19 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | In-crystal reaction cycle of a toluene-bound diiron hydroxylase.
Nature, 544, 2017
|
|
5TDT
 
 | |
5TEN
 
 | | Structure of 4-Hydroxy-tetrahydrodipicolinate Reductase from Vibrio vulnificus with 2,5 Furan Dicarboxylic and NADH with Intact Polyhistidine Tag | | Descriptor: | 2,5 Furan Dicarboxylic Acid, 4-hydroxy-tetrahydrodipicolinate reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Mank, N, Pote, S, Arnette, K, Klapper, V, Chruszcz, M. | | Deposit date: | 2016-09-22 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Comparative structural and mechanistic studies of 4-hydroxy-tetrahydrodipicolinate reductases from Mycobacterium tuberculosis and Vibrio vulnificus.
Biochim Biophys Acta Gen Subj, 1865, 2021
|
|
