3H4Q
 
 | |
3H4W
 
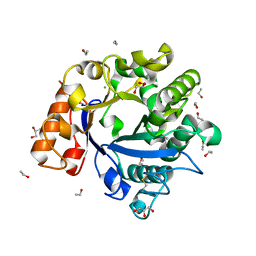 | |
3H6M
 
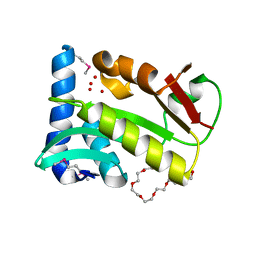
 | | Crystal structure of Staphylococcal nuclease variant Delta+PHS V104E at cryogenic temperature | | Descriptor: | CALCIUM ION, THYMIDINE-3',5'-DIPHOSPHATE, Thermonuclease | | Authors: | Khangulov, V.S, Schlessman, J.L, Heroux, A, Garcia-Moreno, E.B. | | Deposit date: | 2009-04-23 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Staphylococcal nuclease variant Delta+PHS V104E at cryogenic temperature
To be Published
|
|
3H6S
 
 | | Structure of clitocypin - cathepsin V complex | | Descriptor: | Cathepsin L2, Clitocypin analog, SULFATE ION | | Authors: | Renko, M, Sabotic, J, Brzin, J, Turk, D. | | Deposit date: | 2009-04-23 | | Release date: | 2009-10-20 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Versatile loops in mycocypins inhibit three protease families.
J.Biol.Chem., 285, 2010
|
|
3GJX
 
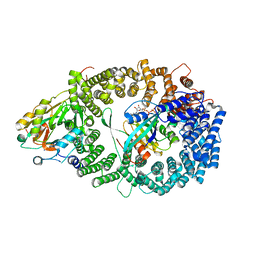 | | Crystal Structure of the Nuclear Export Complex CRM1-Snurportin1-RanGTP | | Descriptor: | CHLORIDE ION, Exportin-1, GTP-binding nuclear protein Ran, ... | | Authors: | Monecke, T, Guettler, T, Neumann, P, Dickmanns, A, Goerlich, D, Ficner, R. | | Deposit date: | 2009-03-09 | | Release date: | 2009-05-26 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Nuclear Export Receptor CRM1 in Complex with Snurportin1 and RanGTP.
Science, 2009
|
|
3H7K
 
 | |
3GMU
 
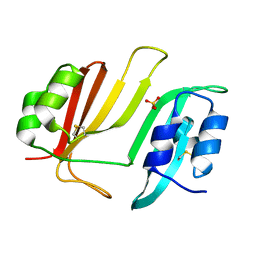 | | Crystal Structure of Beta-Lactamse Inhibitory Protein (BLIP) in Apo Form | | Descriptor: | AMMONIUM ION, Beta-lactamase inhibitory protein, SULFATE ION | | Authors: | Strynadka, N.C.J, Gretes, M, James, M.N.G. | | Deposit date: | 2009-03-15 | | Release date: | 2009-03-31 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Insights into positive and negative requirements for protein-protein interactions by crystallographic analysis of the beta-lactamase inhibitory proteins BLIP, BLIP-I, and BLP.
J.Mol.Biol., 389, 2009
|
|
3GMX
 
 | |
3H8O
 
 | | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked host-guest system | | Descriptor: | 5'-D(*CP*TP*GP*TP*AP*TP*(2YR)P*AP*TP*(MA7)P*GP*CP*G)-3', 5'-D(*TP*CP*GP*CP*TP*AP*TP*AP*AP*TP*AP*CP*A)-3', Alpha-ketoglutarate-dependent dioxygenase alkB homolog 2, ... | | Authors: | Lu, L, Yi, C, Jian, X, Zheng, Q. | | Deposit date: | 2009-04-29 | | Release date: | 2010-03-31 | | Last modified: | 2021-10-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system.
Nucleic Acids Res., 38, 2010
|
|
3GO2
 
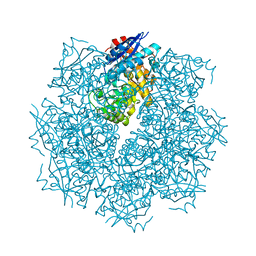 | | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium | | Descriptor: | MAGNESIUM ION, Putative L-alanine-DL-glutamate epimerase | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Chang, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-18 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of putative L-alanine-DL-glutamate epimerase from Burkholderia xenovorans strain LB400 bound to magnesium.
To be Published
|
|
3H94
 
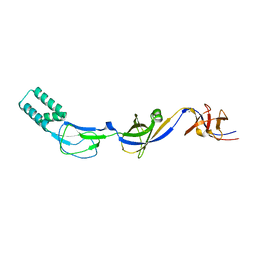 | | Crystal structure of the membrane fusion protein CusB from Escherichia coli | | Descriptor: | Cation efflux system protein cusB, SILVER ION | | Authors: | Su, C.-C, Yang, F, Long, F, Reyon, D, Routh, M.D, Kuo, D.W, Mokhtari, A.K, Van Ornam, J.D, Rabe, K.L, Hoy, J.A, Lee, Y.J, Rajashankar, K.R, Yu, E.W. | | Deposit date: | 2009-04-30 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Crystal structure of the membrane fusion protein CusB from Escherichia coli
J.Mol.Biol., 393, 2009
|
|
3GQL
 
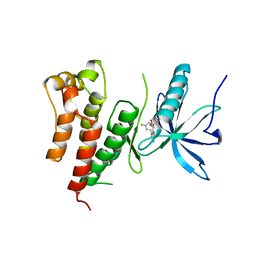 | | Crystal Structure of activated receptor tyrosine kinase in complex with substrates | | Descriptor: | (E)-[4-(3,5-difluorophenyl)-3H-pyrrolo[2,3-b]pyridin-3-ylidene](3-methoxyphenyl)methanol, Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Lew, E.D, Yuzawa, S, Tome, F, Lax, I, Schlessinger, J. | | Deposit date: | 2009-03-24 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site.
Cell(Cambridge,Mass.), 138, 2009
|
|
3GR3
 
 | |
3GRE
 
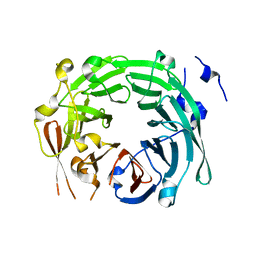 | |
3GRJ
 
 | |
3HAQ
 
 | | Crystal structure of bacteriorhodopsin mutant I148A crystallized from bicelles | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Bacteriorhodopsin, DECANE, ... | | Authors: | Joh, N.H, Yang, D, Bowie, J.U. | | Deposit date: | 2009-05-02 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Similar energetic contributions of packing in the core of membrane and water-soluble proteins.
J.Am.Chem.Soc., 131, 2009
|
|
3HB8
 
 | | Structures of Thymidylate Synthase R163K with Substrates and Inhibitors Show Subunit Asymmetry | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, PHOSPHATE ION, ... | | Authors: | Gibson, L.M, Lovelace, L.L, Lebioda, L. | | Deposit date: | 2009-05-04 | | Release date: | 2010-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structures of human thymidylate synthase R163K with dUMP, FdUMP and glutathione show asymmetric ligand binding.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3GT4
 
 | | Structure of proteinase K with the magic triangle I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, SULFATE ION, proteinase K | | Authors: | Beck, T, Gruene, T, Sheldrick, G.M. | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The magic triangle goes MAD: experimental phasing with a bromine derivative
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3GSX
 
 | | Crystal structure of the binary complex between HLA-A2 and HCMV NLV-T8V peptide variant | | Descriptor: | Beta-2-microglobulin, HCMV pp65 fragment 495-503, variant T8V (NLVPMVAVV), ... | | Authors: | Reiser, J.-B, Saulquin, X, Gras, S, Debeaupuis, E, Echasserieau, K, Kissenpfennig, A, Legoux, F, Chouquet, A, Le Gorrec, M, Machillot, P, Neveu, B, Thielens, N, Malissen, B, Bonneville, M, Housset, D. | | Deposit date: | 2009-03-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural bases for the affinity-driven selection of a public TCR against a dominant human cytomegalovirus epitope.
J.Immunol., 183, 2009
|
|
3GTY
 
 | |
3GVC
 
 | |
3GW0
 
 | | UROD mutant G318R | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3GWU
 
 | | Leucine transporter LeuT in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, LEUCINE, SODIUM ION, ... | | Authors: | Zhou, Z, Zhen, J, Karpowich, N.K, Law, C.J, Reith, M.E.A, Wang, D.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GXT
 
 | |
3GXF
 
 | |
