1UXR
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
6DJ5
 
 | | HIV-1 protease with mutation L76V in complex with GRL-0519 (tris-tetrahydrofuran as P2 ligand) | | Descriptor: | (3R,3aS,3bR,6aS,7aS)-octahydrodifuro[2,3-b:3',2'-d]furan-3-yl [(1S,2R)-1-benzyl-2-hydroxy-3-{[(4-methoxyphenyl)sulfonyl](2-methylpropyl)amino}propyl]carbamate, CHLORIDE ION, GLYCEROL, ... | | Authors: | Wong-Sam, A.E, Wang, Y.F, Weber, I.T. | | Deposit date: | 2018-05-24 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Drug Resistance Mutation L76V Alters Nonpolar Interactions at the Flap-Core Interface of HIV-1 Protease.
ACS Omega, 3, 2018
|
|
1UXT
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | 1-O-phosphono-alpha-D-glucopyranose, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-03-01 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
1V4L
 
 | |
1UMY
 
 | | BHMT from rat liver | | Descriptor: | BETA-MERCAPTOETHANOL, Betaine--homocysteine S-methyltransferase 1, ZINC ION | | Authors: | Gonzalez, B, Pajares, M.A, Sanz-Aparicio, J. | | Deposit date: | 2003-09-02 | | Release date: | 2004-05-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of rat liver betaine homocysteine s-methyltransferase reveals new oligomerization features and conformational changes upon substrate binding.
J. Mol. Biol., 338, 2004
|
|
1M6X
 
 | | Flpe-Holliday Junction Complex | | Descriptor: | Flp recombinase, Symmetrized FRT site | | Authors: | Conway, A.B, Chen, Y, Rice, P.A. | | Deposit date: | 2002-07-17 | | Release date: | 2003-02-04 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Plasticity of the Flp-Holliday Junction Complex
J.Mol.Biol., 326, 2003
|
|
1SF2
 
 | | Structure of E. coli gamma-aminobutyrate aminotransferase | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobutyrate aminotransferase, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Liu, W, Peterson, P.E, Carter, R.J, Zhou, X, Langston, J.A, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-02-19 | | Release date: | 2004-09-14 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of unbound and aminooxyacetate-bound Escherichia coli gamma-aminobutyrate aminotransferase.
Biochemistry, 43, 2004
|
|
1UXN
 
 | | Structural basis for allosteric regulation and substrate specificity of the non-phosphorylating glyceraldehyde-3-phosphate dehydrogenase (GAPN) from Thermoproteus tenax | | Descriptor: | ADENOSINE MONOPHOSPHATE, GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE (NADP+), NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Lorentzen, E, Hensel, R, Pohl, E. | | Deposit date: | 2004-02-27 | | Release date: | 2004-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of Allosteric Regulation and Substrate Specificity of the Non-Phosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Thermoproteus Tenax
J.Mol.Biol., 341, 2004
|
|
4HNX
 
 | | The NatA Acetyltransferase Complex Bound To ppGpp | | Descriptor: | GUANOSINE-5',3'-TETRAPHOSPHATE, N-terminal acetyltransferase A complex catalytic subunit ARD1, N-terminal acetyltransferase A complex subunit NAT1 | | Authors: | Neubauer, J.L, Immormino, R.M, Dollins, D.E, Endo-Streeter, S.T, Pemble IV, C.W, York, J.D. | | Deposit date: | 2012-10-21 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | The Protein Complex NatA Binds Inositol Hexakisphosphate and Exhibits Conformational Flexibility
To be Published
|
|
1RYT
 
 | | RUBRERYTHRIN | | Descriptor: | FE (III) ION, RUBRERYTHRIN | | Authors: | Demare, F, Kurtz, D.M, Nordlund, P. | | Deposit date: | 1996-04-26 | | Release date: | 1997-05-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of Desulfovibrio vulgaris rubrerythrin reveals a unique combination of rubredoxin-like FeS4 and ferritin-like diiron domains.
Nat.Struct.Biol., 3, 1996
|
|
4P3M
 
 | | Crystal structure of serine hydroxymethyltransferase from Psychromonas ingrahamii | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Dworkowski, F, Angelaccio, S, Pascarella, S, Capitani, G. | | Deposit date: | 2014-03-09 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational transitions driven by pyridoxal-5'-phosphate uptake in the psychrophilic serine hydroxymethyltransferase from Psychromonas ingrahamii.
Proteins, 82, 2014
|
|
5ACY
 
 | | N-TERMINAL BROMODOMAIN OF HUMAN BRD4 WITH 1-(2R,4S)-2-methyl-4-(phenylamino)-6-4-(piperidin-1-ylmethyl)phenyl-1,2,3,4- tetrahydroquinolin-1-yl-ethan-1-one | | Descriptor: | 1,2-ETHANEDIOL, 1-[(2S,4R)-2-methyl-4-(phenylamino)-6-[4-(piperidin-1-ylmethyl)phenyl]-3,4-dihydroquinolin-1(2H)-yl]ethanone, BROMODOMAIN-CONTAINING PROTEIN 4 | | Authors: | Chung, C. | | Deposit date: | 2015-08-18 | | Release date: | 2015-10-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Autism-Like Syndrome is Induced by Pharmacological Suppression of Bet Proteins in Young Mice.
J.Exp.Med., 212, 2015
|
|
5SI5
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with N-(12-methoxy-5-methyl-3-propyl-2,4,8,13-tetrazatricyclo[7.4.0.02,6]trideca-1(13),3,5,7,9,11-hexaen-7-yl)methanesulfonamide | | Descriptor: | MAGNESIUM ION, N-[(10S)-2-methoxy-7-methyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazin-6-yl]methanesulfonamide, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SGU
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 12-methoxy-5,7-dimethyl-3-propyl-2,4,8,13-tetrazatricyclo[7.4.0.02,6]trideca-1(13),3,5,7,9,11-hexaene | | Descriptor: | 2-methoxy-6,7-dimethyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
3NTZ
 
 | | Design, Synthesis, Biological Evaluation and X-ray Crystal Structures of Novel Classical 6,5,6-tricyclicbenzo[4,5]thieno[2,3-d]pyrimidines as Dual Thymidylate Synthase and Dihydrofolate Reductase Inhibitors | | Descriptor: | Dihydrofolate reductase, N-[(4-{[(2-amino-4-oxo-3,4-dihydro[1]benzothieno[2,3-d]pyrimidin-5-yl)methyl]amino}phenyl)carbonyl]-L-glutamic acid, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
5SGZ
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 3-(2-chlorophenyl)-12-methoxy-5-methyl-2,4,8,13-tetrazatricyclo[7.4.0.02,6]trideca-1(13),3,5,9,11-pentaen-7-one | | Descriptor: | (10R)-9-(2-chlorophenyl)-2-methoxy-7-methylimidazo[1,5-a]pyrido[3,2-e]pyrazin-6(5H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
2X1X
 
 | | CRYSTAL STRUCTURE OF VEGF-C IN COMPLEX WITH DOMAINS 2 AND 3 OF VEGFR2 IN A TETRAGONAL CRYSTAL FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, ... | | Authors: | Leppanen, V.-M, Prota, A.E, Jeltsch, M, Anisimov, A, Kalkkinen, N, Strandin, T, Lankinen, H, Goldman, A, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-01-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Determinants of Growth Factor Binding and Specificity by Vegf Receptor 2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
7KFA
 
 | | PCSK9 in complex with PCSK9i a 13mer cyclic peptide LDLR disruptor | | Descriptor: | 1-[2,6,10.14-TETRAMETHYL-HEXADECAN-16-YL]-2-[2,10,14-TRIMETHYLHEXADECAN-16-YL]GLYCEROL, CALCIUM ION, Proprotein convertase subtilisin/kexin type 9, ... | | Authors: | Chopra, R, Xu, M, Spraggon, G. | | Deposit date: | 2020-10-13 | | Release date: | 2021-11-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Identification of a PCSK9-LDLR disruptor peptide with in vivo function.
Cell Chem Biol, 29, 2022
|
|
5SHG
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 3-(2-chlorophenyl)-12-methoxy-5-methyl-2,4,8,13-tetrazatricyclo[7.4.0.02,6]trideca-1(9),3,5,7,10,12-hexaen-7-amine | | Descriptor: | (10R)-9-(2-chlorophenyl)-2-methoxy-7-methylimidazo[1,5-a]pyrido[3,2-e]pyrazin-6-amine, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Groebke-Zbinden, K, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
5SJ8
 
 | | Crystal Structure of human phosphodiesterase 10 in complex with 12-methoxy-5-methyl-3-propyl-2,4,8,13-tetrazatricyclo[7.4.0.02,6]trideca-1(13),3,5,7,9,11-hexaene-7-carbonitrile | | Descriptor: | (10S)-2-methoxy-7-methyl-9-propylimidazo[1,5-a]pyrido[3,2-e]pyrazine-6-carbonitrile, MAGNESIUM ION, ZINC ION, ... | | Authors: | Joseph, C, Benz, J, Flohr, A, Rudolph, M.G. | | Deposit date: | 2022-02-01 | | Release date: | 2022-10-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal Structure of a human phosphodiesterase 10 complex
To be published
|
|
7PET
 
 | | The 4x177 nucleosome array containing H1 | | Descriptor: | DNA (702-MER), Histone H1.4, Histone H2A type 1-B/E, ... | | Authors: | Dombrowski, M, Cramer, P. | | Deposit date: | 2021-08-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Histone H1 binding to nucleosome arrays depends on linker DNA length and trajectory.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7OCD
 
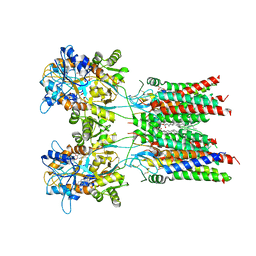 | | Resting state GluA1/A2 heterotetramer in complex with auxiliary subunit TARP gamma 8 (LBD-TMD) | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2, ... | | Authors: | Zhang, D, Watson, J.F, Matthews, P.M, Cais, O, Greger, I.H. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-09 | | Last modified: | 2021-06-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Gating and modulation of a hetero-octameric AMPA glutamate receptor.
Nature, 594, 2021
|
|
3NU0
 
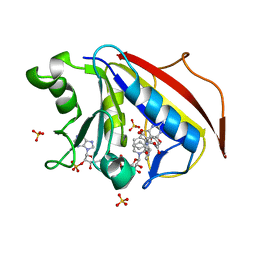 | | Design, Synthesis, Biological Evaluation and X-ray Crystal Structure of Novel Classical 6,5,6-TricyclicBenzo[4,5]thieno[2,3-d]pyrimidines as Dual Thymidylate Synthase and Dihydrofolate Reductase Inhibitors | | Descriptor: | (2S)-2-(5-{[(2-amino-4-oxo-3,4-dihydro[1]benzothieno[2,3-d]pyrimidin-5-yl)methyl]amino}-1-oxo-1,3-dihydro-2H-isoindol-2-yl)pentanedioic acid, Dihydrofolate reducatase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Cody, V. | | Deposit date: | 2010-07-06 | | Release date: | 2011-05-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, synthesis, biological evaluation and X-ray crystal structure of novel classical 6,5,6-tricyclic benzo[4,5]thieno[2,3-d]pyrimidines as dual thymidylate synthase and dihydrofolate reductase inhibitors.
Bioorg.Med.Chem., 19, 2011
|
|
5FJG
 
 | | The crystal structure of light-driven chloride pump ClR in pH 4.5. | | Descriptor: | ANHYDRORETINOL, CHLORIDE ION, CHLORIDE PUMPING RHODOPSIN, ... | | Authors: | Kim, K.L, Kwon, S.K, Jun, S.H, Cha, J.S, Kim, H.Y, Kim, J.H, Cho, H.S. | | Deposit date: | 2015-10-07 | | Release date: | 2016-11-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of Light-Driven Chloride Pump Clr in Ph 4.5.
To be Published
|
|
5GM4
 
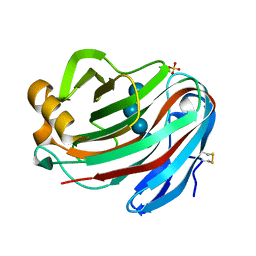 | | Crystal structure of FI-CMCase from Aspergillus aculeatus F-50 in complex with cellotetrose | | Descriptor: | Endoglucanase-1, SULFATE ION, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Huang, J.W, Liu, W.D, Zheng, Y.Y, Chen, C.C, Guo, R.T. | | Deposit date: | 2016-07-12 | | Release date: | 2017-05-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure and genetic modifications of FI-CMCase from Aspergillus aculeatus F-50
Biochem. Biophys. Res. Commun., 478, 2016
|
|
