8WQJ
 
 | |
8WQI
 
 | |
8WQH
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQG
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.09 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQF
 
 | | cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 2) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQE
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CUX1 (conformation 1) | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQD
 
 | |
8WQC
 
 | | cryo-EM structure of neddylated CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CDK5R1 | | Descriptor: | Cullin-2, E3 ubiquitin-protein ligase RBX1, N-terminally processed, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQB
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 2) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQA
 
 | | Cryo-EM structure of CUL2-RBX1-ELOB-ELOC-FEM1B bound with the C-degron of CCDC89 (conformation 1) | | Descriptor: | Coiled-coil domain-containing protein 89, Cullin-2, E3 ubiquitin-protein ligase RBX1, ... | | Authors: | Chen, X, Zhang, K, Xu, C. | | Deposit date: | 2023-10-11 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Mechanism of Psi-Pro/C-degron recognition by the CRL2 FEM1B ubiquitin ligase.
Nat Commun, 15, 2024
|
|
8WQ9
 
 | |
8WQ8
 
 | |
8WQ7
 
 | | Crystal structure of d(CGTATACG)2 with a four-carbon linker containing diacridine compound | | Descriptor: | DNA (5'-D(P*CP*GP*TP*AP*TP*AP*CP*G)-3'), MANGANESE (II) ION, N,N'-di(acridin-9-yl)butane-1,4-diamine | | Authors: | Huang, S.C, Satange, R.B, Hou, M.H. | | Deposit date: | 2023-10-11 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Targeting DNA junction sites by bis-intercalators induces topological changes with potent antitumor effects.
Nucleic Acids Res., 2024
|
|
8WQ4
 
 | |
8WQ2
 
 | |
8WQ0
 
 | | Cryo-EM structure of WIV1 spike glycoprotein (the closed state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Wang, X, Qiao, S. | | Deposit date: | 2023-10-10 | | Release date: | 2024-07-17 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Structural determinants of spike infectivity in bat SARS-like coronaviruses RsSHC014 and WIV1.
J.Virol., 2024
|
|
8WPZ
 
 | |
8WPV
 
 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum soaked in Fe2+ solution for 30min | | Descriptor: | CHLORIDE ION, FE (III) ION, MAGNESIUM ION, ... | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.059 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WPU
 
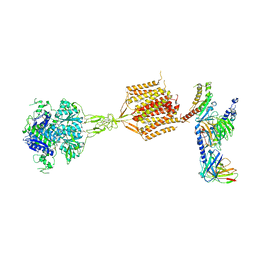 | | Human calcium-sensing receptor(CaSR) bound to cinacalcet in complex with Gq protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPT
 
 | | Truncated mutant (1-171) of ferritin from Ureaplasma diversum | | Descriptor: | CHLORIDE ION, FE (III) ION, Truncated ferritin | | Authors: | Wang, W.M, Xi, H.F, Gong, W.J, Ma, D.Y, Wang, H.F. | | Deposit date: | 2023-10-10 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Growth Process of Fe-O Nanoclusters with Different Sizes Biosynthesized by Protein Nanocages.
J.Am.Chem.Soc., 146, 2024
|
|
8WPP
 
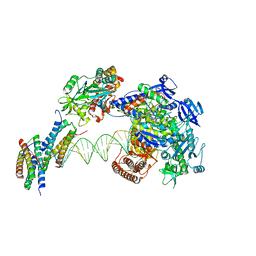 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with endogenous DNA | | Descriptor: | A22R DNA polymerase processivity factor, DNA polymerase, E4R Uracil-DNA glycosylase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPK
 
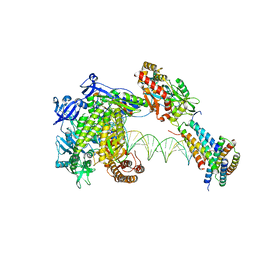 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exgenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, DNA polymerase, DNA polymerase processivity factor, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPG
 
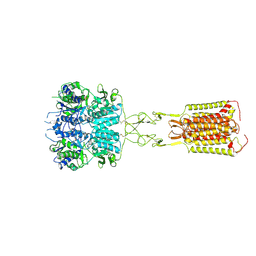 | | Human calcium-sensing receptor bound with cinacalcet in detergent | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Ling, S.L, Meng, X.Y, Tian, C.L. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into asymmetric activation of the calcium-sensing receptor-G q complex.
Cell Res., 34, 2024
|
|
8WPF
 
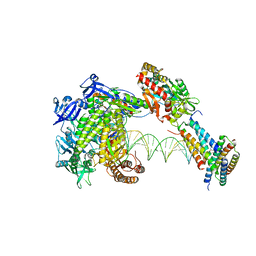 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 with exogenous DNA bearing one abasic site | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
8WPE
 
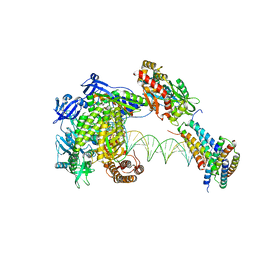 | | Structure of monkeypox virus polymerase complex F8-A22-E4-H5 (tag-free A22) with exogenous DNA | | Descriptor: | 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, A22R DNA polymerase processivity factor, DNA polymerase, ... | | Authors: | Wang, X, Li, N, Gao, N. | | Deposit date: | 2023-10-10 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural insights into the assembly and mechanism of mpox virus DNA polymerase complex F8-A22-E4-H5.
Mol.Cell, 83, 2023
|
|
