3AOI
 
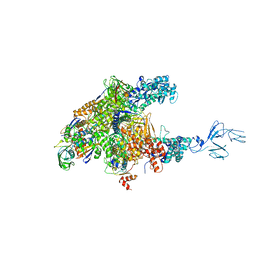 | | RNA polymerase-Gfh1 complex (Crystal type 2) | | Descriptor: | Anti-cleavage anti-GreA transcription factor Gfh1, DNA (5'-D(*GP*GP*TP*CP*TP*GP*TP*AP*TP*CP*AP*CP*GP*AP*GP*CP*CP*A*CP*CP*GP*CP*CP*GP*CP*AP*T)-3'), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Tagami, S, Sekine, S, Kumarevel, T, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-09-30 | | Release date: | 2010-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Crystal structure of bacterial RNA polymerase bound with a transcription inhibitor protein
Nature, 468, 2010
|
|
2ZZP
 
 | |
337D
 
 | |
349D
 
 | |
3A5T
 
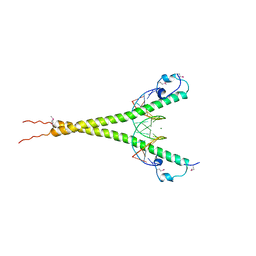 | | Crystal structure of MafG-DNA complex | | Descriptor: | 5'-D(*CP*TP*GP*AP*TP*GP*AP*GP*TP*CP*AP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*TP*GP*AP*CP*TP*CP*AP*TP*CP*AP*G)-3', MAGNESIUM ION, ... | | Authors: | Kurokawa, H, Motohashi, H, Sueno, S, Kimura, M, Takagawa, H, Kanno, Y, Yamamoto, M, Tanaka, T. | | Deposit date: | 2009-08-11 | | Release date: | 2009-10-13 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Basis of Alternative DNA Recognition by Maf Transcription Factors
Mol.Cell.Biol., 29, 2009
|
|
3A62
 
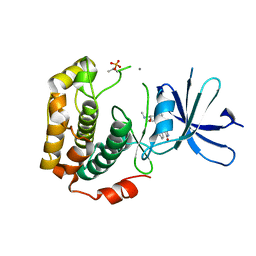 | | Crystal structure of phosphorylated p70S6K1 | | Descriptor: | MANGANESE (II) ION, Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A2F
 
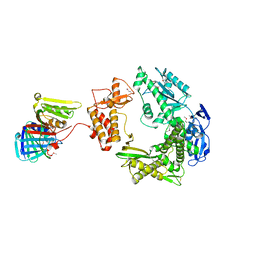 | |
3A61
 
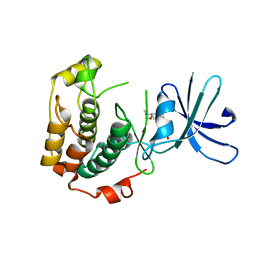 | | Crystal structure of unphosphorylated p70S6K1 (Form II) | | Descriptor: | Ribosomal protein S6 kinase beta-1, STAUROSPORINE | | Authors: | Sunami, T, Byrne, N, Diehl, R.E, Funabashi, K, Hall, D.L, Ikuta, M, Patel, S.B, Shipman, J.M, Smith, R.F, Takahashi, I, Zugay-Murphy, J, Iwasawa, Y, Lumb, K.J, Munshi, S.K, Sharma, S. | | Deposit date: | 2009-08-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Structural basis of human p70 ribosomal S6 kinase-1 regulation by activation loop phosphorylation.
J.Biol.Chem., 285, 2010
|
|
3AF6
 
 | | The crystal structure of an archaeal CPSF subunit, PH1404 from Pyrococcus horikoshii complexed with RNA-analog | | Descriptor: | 5'-R(*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU)P*(SSU))-3', Putative uncharacterized protein PH1404, SULFATE ION, ... | | Authors: | Nishida, Y, Ishikawa, H, Nakagawa, N, Masui, R, Kuramitsu, S. | | Deposit date: | 2010-02-24 | | Release date: | 2010-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an archaeal cleavage and polyadenylation specificity factor subunit from Pyrococcus horikoshii
Proteins, 78, 2010
|
|
3CR9
 
 | | Crystal structure of the complex of Lactoferrin with 6-(Hydroxymethyl)oxane-2,3,4,5-tetrol at 3.49 A resolution | | Descriptor: | FE (III) ION, Lactotransferrin, alpha-D-glucopyranose | | Authors: | Mir, R, Kaur, A, Singh, A.K, Singh, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2008-04-05 | | Release date: | 2008-04-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.49 Å) | | Cite: | Crystal structure of the complex of Lactoferrin with 6-(Hydroxymethyl)oxane-2,3,4,5-tetrol at 3.49 A resolution
To be Published
|
|
3CRX
 
 | |
3CHP
 
 | | Crystal structure of leukotriene a4 hydrolase in complex with (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid | | Descriptor: | (3S)-3-amino-4-oxo-4-[(4-phenylmethoxyphenyl)amino]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Thunnissen, M.M.G.M, Adler, M, Whitlow, M. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Synthesis of glutamic acid analogs as potent inhibitors of leukotriene A4 hydrolase.
Bioorg.Med.Chem., 16, 2008
|
|
3CTF
 
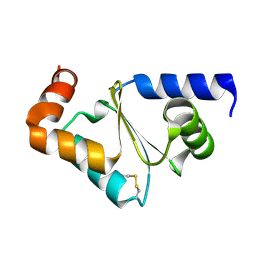 | | Crystal structure of oxidized GRX2 | | Descriptor: | Glutaredoxin-2 | | Authors: | Yu, J, Teng, Y.B, Zhou, C.Z. | | Deposit date: | 2008-04-14 | | Release date: | 2008-11-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the different activities of yeast Grx1 and Grx2.
Biochim.Biophys.Acta, 1804, 2010
|
|
3COB
 
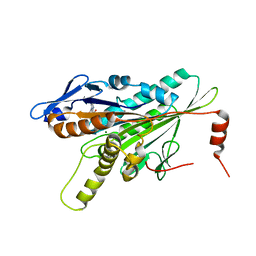 | | Structural Dynamics of the Microtubule binding and regulatory elements in the Kinesin-like Calmodulin binding protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin heavy chain-like protein, MAGNESIUM ION | | Authors: | Vinogradova, M.V, Malanina, G.G, Reddy, V, Reddy, A.S.N, Fletterick, R.J. | | Deposit date: | 2008-03-27 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural dynamics of the microtubule binding and regulatory elements in the kinesin-like calmodulin binding protein.
J.Struct.Biol., 163, 2008
|
|
3CPU
 
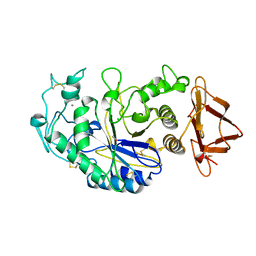 | | SUBSITE MAPPING OF THE ACTIVE SITE OF HUMAN PANCREATIC ALPHA-AMYLASE USING SUBSTRATES, THE PHARMACOLOGICAL INHIBITOR ACARBOSE, AND AN ACTIVE SITE VARIANT | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pancreatic alpha-amylase, ... | | Authors: | Brayer, G.D, Sidhu, G, Maurus, R, Rydberg, E.H, Braun, C, Wang, Y, Nguyen, N.T, Overall, C.M, Withers, S.G. | | Deposit date: | 1999-06-08 | | Release date: | 2001-06-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subsite mapping of the human pancreatic alpha-amylase active site through structural, kinetic, and mutagenesis techniques.
Biochemistry, 39, 2000
|
|
3CQO
 
 | |
3CXF
 
 | | Crystal structure of transthyretin variant Y114H | | Descriptor: | Transthyretin | | Authors: | Cendron, L, Zanotti, G, Folli, C, Alfieri, B, Pasquato, N, Berni, R. | | Deposit date: | 2008-04-24 | | Release date: | 2009-04-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mutational analyses of protein-protein interactions between transthyretin and retinol-binding protein.
Febs J., 275, 2008
|
|
3CTS
 
 | |
3CN9
 
 | |
3CS7
 
 | | FACTOR XA IN COMPLEX WITH THE INHIBITOR 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one | | Descriptor: | 1-(4-methoxyphenyl)-6-(4-(1-(pyrrolidin-1-ylmethyl)cyclopropyl)phenyl)-3-(trifluoromethyl)-5,6-dihydro-1H-pyrazolo[3,4-c]pyridin-7(4H)-one, CALCIUM ION, Coagulation factor X | | Authors: | Alexander, R.S. | | Deposit date: | 2008-04-09 | | Release date: | 2008-07-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Achieving structural diversity using the perpendicular conformation of alpha-substituted phenylcyclopropanes to mimic the bioactive conformation of ortho-substituted biphenyl P4 moieties: discovery of novel, highly potent inhibitors of Factor Xa.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D5N
 
 | | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125. | | Descriptor: | Q97W15_SULSO | | Authors: | Vorobiev, S.M, Chen, Y, Seetharaman, J, Lee, D, Foote, R.E, Maglaqui, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-05-16 | | Release date: | 2008-07-15 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the Q97W15_SULSO protein from Sulfolobus solfataricus. NESG target SsR125.
To be Published
|
|
3D6Y
 
 | |
3D4J
 
 | | Crystal structure of Human mevalonate diphosphate decarboxylase | | Descriptor: | Diphosphomevalonate decarboxylase, SULFATE ION | | Authors: | Voynova, N.E, Fu, Z, Battaile, K, Herdendorf, T.J, Kim, J.-J.P, Miziorko, H.M. | | Deposit date: | 2008-05-14 | | Release date: | 2008-12-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human mevalonate diphosphate decarboxylase: characterization, investigation of the mevalonate diphosphate binding site, and crystal structure.
Arch.Biochem.Biophys., 480, 2008
|
|
3CGW
 
 | | Crystal structure of 2-phospho-(S)-lactate transferase from Methanosarcina mazei. Northeast Structural Genomics Consortium target MaR46 | | Descriptor: | LPPG:FO 2-phospho-L-lactate transferase | | Authors: | Forouhar, F, Abashidze, M, Seetharaman, J, Vorobiev, S.M, Ciao, M, Janjua, H, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-03-06 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Molecular insights into the biosynthesis of the f420 coenzyme.
J.Biol.Chem., 283, 2008
|
|
