1EGL
 
 | |
5T5N
 
 | | Calcium-activated chloride channel bestrophin-1 (BEST1), triple mutant: I76A, F80A, F84A; in complex with an Fab antibody fragment, chloride, and calcium | | Descriptor: | CALCIUM ION, CHLORIDE ION, Fab antibody fragment, ... | | Authors: | Long, S.B, Vaisey, G. | | Deposit date: | 2016-08-31 | | Release date: | 2016-11-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Distinct regions that control ion selectivity and calcium-dependent activation in the bestrophin ion channel.
Proc. Natl. Acad. Sci. U.S.A., 113, 2016
|
|
8I5B
 
 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
6DJH
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ515 | | Descriptor: | 1,2-ETHANEDIOL, 8-bromo-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.918 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
5DWJ
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain in Complex with a Resorcinyl 4-Fluoro-substituted Diaryl-imine analog 4-[(E)-[(4-fluorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol | | Descriptor: | 4-[(E)-[(4-fluorophenyl)imino](4-hydroxyphenyl)methyl]benzene-1,3-diol, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Wright, N.J, Srinivasan, S, Zheng, Y, Wang, S, Min, J, Dong, C, Liao, Z, Cavett, V, Nowak, J, Houtman, R, Carlson, K.E, Josan, J.S, Elemento, O, Katzenellenbogen, J.A, Zhou, H.B, Nettles, K.W. | | Deposit date: | 2015-09-22 | | Release date: | 2016-05-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Predictive features of ligand-specific signaling through the estrogen receptor.
Mol.Syst.Biol., 12, 2016
|
|
6ME1
 
 | |
6RMF
 
 | | Crystal structure of NDM-1 with VNRX-5133 | | Descriptor: | (10aR)-2-(((1r,4R)-4-((2-aminoethyl)amino)cyclohexyl)methyl)-6-carboxy-4-hydroxy-4,10a-dihydro-10H-benzo[5,6][1,2]oxaborinino[2,3-b][1,4,2]oxazaborol-4-uide, (4~{R})-4-[2-[4-(2-azanylethylamino)cyclohexyl]ethanoylamino]-3,3-bis(oxidanyl)-2-oxa-3-boranuidabicyclo[4.4.0]deca-1(10),6,8-triene-10-carboxylic acid, Metallo-beta-lactamase type 2, ... | | Authors: | Hinchliffe, P, Spencer, J, Brem, J, Schofield, C.J. | | Deposit date: | 2019-05-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Bicyclic Boronate VNRX-5133 Inhibits Metallo- and Serine-beta-Lactamases.
J.Med.Chem., 62, 2019
|
|
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5TXU
 
 | | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Nocadello, S, Minasov, G, Kiryukhina, O, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation
To Be Published
|
|
6FXJ
 
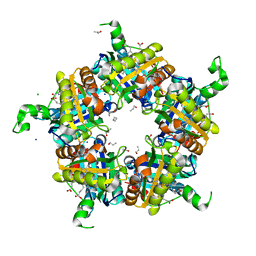 | | Structure of coproheme decarboxylase from Listeria monocytogenes in complex with iron coproporphyrin III | | Descriptor: | 1,3,5,8-TETRAMETHYL-PORPHINE-2,4,6,7-TETRAPROPIONIC ACID FERROUS COMPLEX, CHLORIDE ION, N-PROPANOL, ... | | Authors: | Hofbauer, S, Pfanzagl, V, Mlynek, G. | | Deposit date: | 2018-03-09 | | Release date: | 2019-07-10 | | Last modified: | 2025-10-01 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Redox Cofactor Rotates during Its Stepwise Decarboxylation: Molecular Mechanism of Conversion of Coproheme to Hemeb.
Acs Catalysis, 9, 2019
|
|
6BYW
 
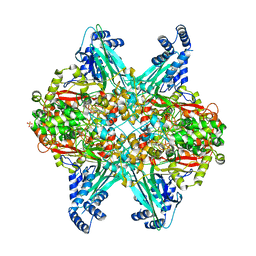 | | Structure of GoxA from Pseudoalteromonas luteoviolacea | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GoxA, ... | | Authors: | Yukl, E.T, Avalos, D. | | Deposit date: | 2017-12-21 | | Release date: | 2018-02-14 | | Last modified: | 2025-04-02 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and Enzymatic Properties of an Unusual Cysteine Tryptophylquinone-Dependent Glycine Oxidase from Pseudoalteromonas luteoviolacea.
Biochemistry, 57, 2018
|
|
5CID
 
 | |
6MTU
 
 | | Crystal structure of human Scribble PDZ1:pMCC complex | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Colorectal mutant cancer protein, ... | | Authors: | Caria, S, Stewart, B.Z, Humbert, P.O, Kvansakul, M. | | Deposit date: | 2018-10-22 | | Release date: | 2019-08-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.141 Å) | | Cite: | Structural analysis of phosphorylation-associated interactions of human MCC with Scribble PDZ domains.
Febs J., 286, 2019
|
|
5L63
 
 | | Yeast 20S proteasome with human beta5c (1-138) and human beta6 (97-111; 118-133) in complex with epoxyketone inhibitor 17 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Groll, M, Huber, E.M. | | Deposit date: | 2016-05-28 | | Release date: | 2016-11-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A humanized yeast proteasome identifies unique binding modes of inhibitors for the immunosubunit beta 5i.
EMBO J., 35, 2016
|
|
6MUQ
 
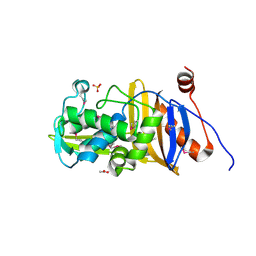 | | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica. | | Descriptor: | ACETATE ION, Murein-DD-endopeptidase, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-10-23 | | Release date: | 2018-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | 1.67 Angstrom Resolution Crystal Structure of Murein-DD-endopeptidase from Yersinia enterocolitica.
To Be Published
|
|
6DJJ
 
 | | Crystal structure of Tdp1 catalytic domain in complex with compound XZ532 | | Descriptor: | 1,2-ETHANEDIOL, 4-aminobenzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Burke Jr, T.R, Pommier, Y, Waugh, D.S. | | Deposit date: | 2018-05-25 | | Release date: | 2019-05-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.741 Å) | | Cite: | Identification of a ligand binding hot spot and structural motifs replicating aspects of tyrosyl-DNA phosphodiesterase I (TDP1) phosphoryl recognition by crystallographic fragment cocktail screening.
Nucleic Acids Res., 47, 2019
|
|
5K6W
 
 | | Sidekick-1 immunoglobulin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Goodman, K.M, Mannepalli, S, Honig, B, Shapiro, L. | | Deposit date: | 2016-05-25 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Molecular basis of sidekick-mediated cell-cell adhesion and specificity.
Elife, 5, 2016
|
|
8XYG
 
 | | Crystal structure of SARS-CoV-2 BQ.1.1 RBD and human ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Lan, J, Wang, C.H. | | Deposit date: | 2024-01-19 | | Release date: | 2025-01-22 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Receptor binding mechanism and immune evasion capacity of SARS-CoV-2 BQ.1.1 lineage.
Virology, 600, 2024
|
|
6N3S
 
 | | Crystal structure of apo-cruzain | | Descriptor: | 1,2-ETHANEDIOL, Cruzipain, PHOSPHATE ION | | Authors: | Silva, E.B, Dall, E, Rodrigues, F.T.G, Ferreira, R.S, Brandstetter, H. | | Deposit date: | 2018-11-16 | | Release date: | 2019-05-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.193 Å) | | Cite: | Cruzain structures: apocruzain and cruzain bound to S-methyl thiomethanesulfonate and implications for drug design.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
9JAK
 
 | | P[28] rotavirus VP8* protein with LNFP-I | | Descriptor: | Outer capsid protein VP4, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Zheng, Y, Yan, J. | | Deposit date: | 2024-08-24 | | Release date: | 2025-07-09 | | Last modified: | 2025-10-22 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Specific binding of human P[28] rotavirus VP8* protein to blood group ABH antigens on type 1 chains.
Plos Pathog., 21, 2025
|
|
5C3J
 
 | | Crystal structure of Mitochondrial rhodoquinol-fumarate reductase from Ascaris suum with Ubiquinone-1 | | Descriptor: | Cytochrome b-large subunit, FE2/S2 (INORGANIC) CLUSTER, FE3-S4 CLUSTER, ... | | Authors: | Harada, S, Shiba, T, Sato, D, Yamamoto, A, Nagahama, M, Yone, A, Inaoka, D.K, Sakamoto, K, Inoue, M, Honma, T, Kita, K. | | Deposit date: | 2015-06-17 | | Release date: | 2016-06-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
To Be Published
|
|
7KOS
 
 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | Descriptor: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2020-11-09 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
6BUQ
 
 | |
5UME
 
 | | Crystal Structure of 5,10-Methylenetetrahydrofolate Reductase MetF from Haemophilus influenzae | | Descriptor: | 1,2-ETHANEDIOL, 5,10-methylenetetrahydrofolate reductase, ACETIC ACID, ... | | Authors: | Kim, Y, Mulligan, R, Maltseva, N, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-27 | | Release date: | 2017-02-22 | | Last modified: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural genomics of bacterial drug targets: Application of a high-throughput pipeline to solve 58 protein structures from pathogenic and related bacteria.
Microbiol Resour Announc, 14, 2025
|
|
5UJF
 
 | |
