1VJC
 
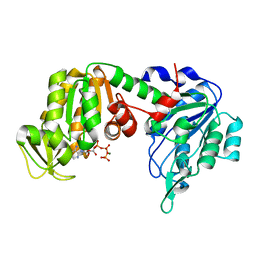 | | Structure of pig muscle PGK complexed with MgATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, phosphoglycerate kinase | | Authors: | Flachner, B, Kovari, Z, Varga, A, Gugolya, Z, Vonderviszt, F, Naray-Szabo, G, Vas, M. | | Deposit date: | 2004-02-03 | | Release date: | 2004-03-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Role of phosphate chain mobility of MgATP in completing the 3-phosphoglycerate kinase catalytic site: binding, kinetic, and crystallographic studies with ATP and MgATP.
Biochemistry, 43, 2004
|
|
3L7I
 
 | |
2VY1
 
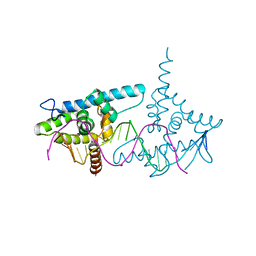 | | Structure of LEAFY transcription factor from Arabidopsis thaliana in complex with DNA from AP1 promoter | | Descriptor: | 5'-D(*TP*TP*AP*CP*GP*GP*AP*CP*CP*AP *CP*TP*GP*GP*TP*CP*CP*TP*TP*CP)-3', PROTEIN LEAFY | | Authors: | Hames, C, Ptchelkine, D, Grimm, C, Thevenon, E, Moyroud, E, Gerard, F, Martiel, J.L, Benlloch, R, Parcy, F, Muller, C.W. | | Deposit date: | 2008-07-16 | | Release date: | 2008-09-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.104 Å) | | Cite: | Structural Basis for Leafy Floral Switch Function and Similarity with Helix-Turn-Helix Proteins.
Embo J., 27, 2008
|
|
3L92
 
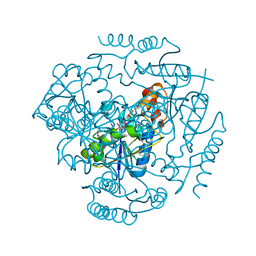 | | Phosphopantetheine adenylyltransferase from Yersinia pestis complexed with coenzyme A. | | Descriptor: | COENZYME A, Phosphopantetheine adenylyltransferase | | Authors: | Osipiuk, J, Maltseva, N, Makowska-grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-01-04 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | X-ray crystal structure of phosphopantetheine adenylyltransferase from Yersinia pestis.
To be Published
|
|
6GSU
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
1VRK
 
 | |
6GSW
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6GSX
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | L-gamma-glutamyl-S-[(9S,10S)-10-hydroxy-9,10-dihydrophenanthren-9-yl]-L-cysteinylglycine, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3, SULFATE ION | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
2VO2
 
 | | Crystal structure of soybean ascorbate peroxidase mutant W41A subjected to low dose X-rays | | Descriptor: | ASCORBATE PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Metcalfe, C.L, Badyal, S.K, Raven, E.L, Moody, P.C.E. | | Deposit date: | 2008-02-08 | | Release date: | 2008-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Iron Oxidation State Modulates Active Site Structure in a Heme Peroxidase.
Biochemistry, 47, 2008
|
|
3AFF
 
 | | Crystal structure of the HsaA monooxygenase from M. tuberculosis | | Descriptor: | Hydroxylase, putative | | Authors: | D'Angelo, I, Lin, L.Y, Dresen, C, Tocheva, E.I, Strynadka, N, Eltis, L.D. | | Deposit date: | 2010-02-28 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A flavin-dependent monooxygenase from Mycobacterium tuberculosis involved in cholesterol catabolism
J.Biol.Chem., 285, 2010
|
|
6GST
 
 | | FIRST-SPHERE AND SECOND-SPHERE ELECTROSTATIC EFFECTS IN THE ACTIVE SITE OF A CLASS MU GLUTATHIONE TRANSFERASE | | Descriptor: | GLUTATHIONE, MU CLASS GLUTATHIONE S-TRANSFERASE OF ISOENZYME 3-3 | | Authors: | Xiao, G, Ji, X, Armstrong, R.N, Gilliland, G.L. | | Deposit date: | 1996-01-26 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | First-sphere and second-sphere electrostatic effects in the active site of a class mu gluthathione transferase.
Biochemistry, 35, 1996
|
|
6H90
 
 | |
1VRL
 
 | | MutY adenine glycosylase in complex with DNA and soaked adenine free base | | Descriptor: | 5'-D(*AP*AP*GP*AP*CP*(8OG)P*TP*GP*GP*AP*C)-3', 5'-D(*TP*GP*TP*CP*CP*AP*(HPD)P*GP*TP*CP*T)-3', ADENINE, ... | | Authors: | Fromme, J.C, Banerjee, A, Huang, S.J, Verdine, G.L. | | Deposit date: | 2005-03-08 | | Release date: | 2005-03-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for removal of adenine mispaired with 8-oxoguanine by MutY adenine DNA glycosylase
Nature, 427, 2004
|
|
2VUZ
 
 | | Crystal structure of Codakine in complex with biantennary nonasaccharide at 1.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CODAKINE, ... | | Authors: | Gourdine, J.P, Cioci, G.C, Miguet, L, Unverzagt, C, Varrot, A, Gauthier, C, Smith-Ravin, E.J, Imberty, A. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High Affinity Interaction between a Bivalve C-Type Lectin and a Biantennary Complex-Type N-Glycan Revealed by Crystallography and Microcalorimetry.
J.Biol.Chem., 283, 2008
|
|
2WFA
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Beryllium trifluoride, in an open conformation. | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Near Attack Conformers Dominate Beta-Phosphoglucomutase Complexes Where Geometry and Charge Distribution Reflect Those of Substrate.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2ZXG
 
 | | Aminopeptidase N complexed with the aminophosphinic inhibitor of PL250, a transition state analogue | | Descriptor: | Aminopeptidase N, GLYCEROL, N-{(2S)-3-[(1R)-1-aminoethyl](hydroxy)phosphoryl-2-benzylpropanoyl}-L-phenylalanine, ... | | Authors: | Nakajima, Y, Ito, K, Yoshimoto, T. | | Deposit date: | 2008-12-24 | | Release date: | 2009-08-25 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structure of aminopeptidase N from Escherichia coli complexed with the transition-state analogue aminophosphinic inhibitor PL250
Acta Crystallogr.,Sect.D, 65, 2009
|
|
8Q7Y
 
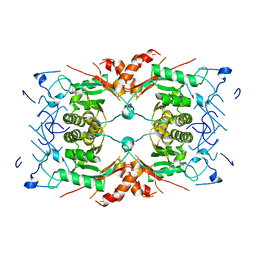 | |
6H91
 
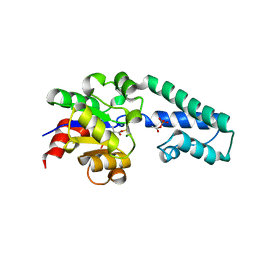 | |
2VZ3
 
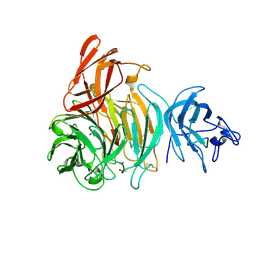 | | bleached galactose oxidase | | Descriptor: | ACETATE ION, COPPER (II) ION, GALACTOSE OXIDASE | | Authors: | Rogers, M.S, Hurtado-Guerrero, R, Firbank, S.J, Halcrow, M.A, Dooley, D.M, Phillips, S.E.V, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2008-07-29 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cross-Link Formation of the Cysteine 228-Tyrosine 272 Catalytic Cofactor of Galactose Oxidase Does not Require Dioxygen.
Biochemistry, 47, 2008
|
|
2XQ2
 
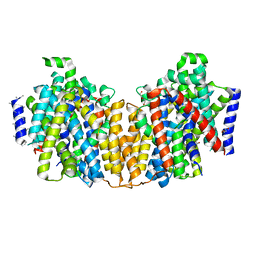 | | Structure of the K294A mutant of vSGLT | | Descriptor: | DI(HYDROXYETHYL)ETHER, SODIUM/GLUCOSE COTRANSPORTER | | Authors: | Watanabe, A, Choe, S, Chaptal, V, Rosenberg, J.M, Wright, E.M, Grabe, M, Abramson, J. | | Deposit date: | 2010-09-01 | | Release date: | 2010-12-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | The Mechanism of Sodium and Substrate Release from the Binding Pocket of Vsglt
Nature, 468, 2010
|
|
3LLQ
 
 | |
2VUJ
 
 | | Environmentally isolated GH11 xylanase | | Descriptor: | GH11 XYLANASE, GLYCEROL | | Authors: | Dumon, C, Varvak, A, Wall, M.A, Flint, J.E, Lewis, R.J, Lakey, J.H, Luginbuhl, P, Healey, S, Todaro, T, DeSantis, G, Sun, M, Parra-Gessert, L, Tan, X, Weiner, D.P, Gilbert, H.J. | | Deposit date: | 2008-05-26 | | Release date: | 2008-06-17 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Engineering Hyperthermostability Into a Gh11 Xylanase is Mediated by Subtle Changes to Protein Structure.
J.Biol.Chem., 283, 2008
|
|
2VS5
 
 | | THE BINDING OF UDP-GALACTOSE BY AN ACTIVE SITE MUTANT OF alpha-1,3 GALACTOSYLTRANSFERASE (alpha3GT) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GALACTOSE-URIDINE-5'-DIPHOSPHATE, MANGANESE (II) ION, ... | | Authors: | Tumbale, P, Jamaluddin, H, Thiyagarajan, N, Brew, K, Acharya, K.R. | | Deposit date: | 2008-04-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structural Basis of Udp-Galactose Binding by Alpha- 1,3-Galactosyltransferase (Alpha3Gt): Role of Negative Charge on Aspartic Acid 316 in Structure and Activity.
Biochemistry, 47, 2008
|
|
1W4F
 
 | | Peripheral-subunit from mesophilic, thermophilic and hyperthermophilic bacteria fold by ultrafast, apparently two-state transitions | | Descriptor: | DIHYDROLIPOYLLYSINE-RESIDUE ACETYLTRANSFERASE | | Authors: | Ferguson, N, Sharpe, T.D, Schartau, P.J, Allen, M.D, Johnson, C.M, Fersht, A.R. | | Deposit date: | 2004-07-23 | | Release date: | 2005-07-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Ultra-Fast Barrier-Limited Folding in the Peripheral Subunit-Binding Domain Family.
J.Mol.Biol., 353, 2005
|
|
2XL7
 
 | | Structure and metal-loading of a soluble periplasm cupro-protein: Cu- CucA-closed (SeMet) | | Descriptor: | CHLORIDE ION, COPPER (II) ION, SLL1785 PROTEIN, ... | | Authors: | Waldron, K.J, Firbank, S.J, Dainty, S.J, Perez-Rama, M, Tottey, S, Robinson, N.J. | | Deposit date: | 2010-07-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and Metal Loading of a Soluble Periplasm Cuproprotein.
J.Biol.Chem., 285, 2010
|
|
