5L0K
 
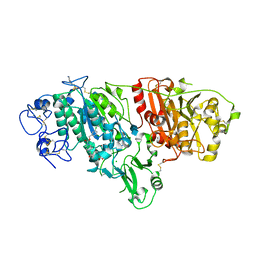 | | Crystal Structure of Autotaxin and Compound PF-8380 | | Descriptor: | (3,5-dichlorophenyl)methyl 4-[3-oxo-3-(2-oxo-2,3-dihydro-1,3-benzoxazol-6-yl)propyl]piperazine-1-carboxylate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Durbin, J.D. | | Deposit date: | 2016-07-27 | | Release date: | 2016-08-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Novel Autotaxin Inhibitors for the Treatment of Osteoarthritis Pain: Lead Optimization via Structure-Based Drug Design.
Acs Med.Chem.Lett., 7, 2016
|
|
4UCG
 
 | | NmeDAH7PS R126S variant | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, PHOSPHO-2-DEHYDRO-3-DEOXYHEPTONATE ALDOLASE, ... | | Authors: | Cross, P.J, Heyes, L.C, Zhang, S, Nazmi, A.R, Parker, E.J. | | Deposit date: | 2014-12-03 | | Release date: | 2016-01-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Functional Unit of Neisseria Meningitidis 3-Deoxy-D-Arabino-Heptulosonate 7-Phosphate Synthase is Dimeric.
Plos One, 11, 2016
|
|
4UDC
 
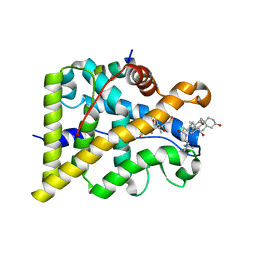 | | GR in complex with dexamethasone | | Descriptor: | 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, DEXAMETHASONE, GLUCOCORTICOID RECEPTOR, ... | | Authors: | Edman, K, Hogner, A, Hussein, A, Bjursell, M, Aagaard, A, Backstrom, S, Bodin, C, Wissler, L, Jellesmark-Jensen, T, Cavallin, A, Karlsson, U, Nilsson, E, Lecina, D, Takahashi, R, Grebner, C, Lepisto, M, Guallar, V. | | Deposit date: | 2014-12-09 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Binding Mechanism in Steroid Receptors: From Conserved Plasticity to Differential Evolutionary Constraints.
Structure, 23, 2015
|
|
4UCP
 
 | |
4UDN
 
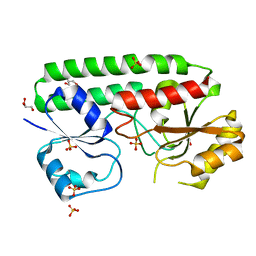 | | structure of metal-free periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2014-12-10 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal Structure of a Periplasmic Solute Binding Protein in Metal-Free, Intermediate and Metal-Bound States from Candidatus Liberibacter Asiaticus.
J.Struct.Biol., 189, 2015
|
|
4UED
 
 | |
4UEO
 
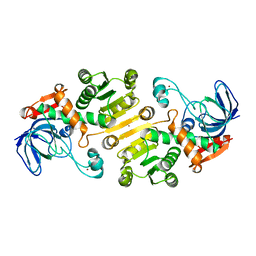 | | Open state of galactitol-1-phosphate 5-dehydrogenase from E. coli, with zinc in the catalytic site. | | Descriptor: | GALACTITOL-1-PHOSPHATE 5-DEHYDROGENASE, ZINC ION | | Authors: | Benavente, R, Esteban-Torres, M, Kohring, G.W, Cortes-Cabrera, A, Gago, F, Acebron, I, de las Rivas, B, Munoz, R, Mancheno, J.M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantioselective Oxidation of Galactitol 1-Phosphate by Galactitol-1-Phosphate 5-Dehydrogenase from Escherichia Coli
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4UEM
 
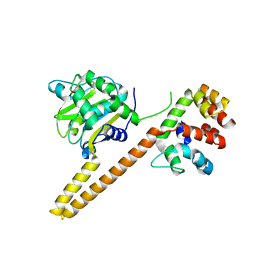 | | UCH-L5 in complex with the RPN13 DEUBAD domain | | Descriptor: | PROTEASOMAL UBIQUITIN RECEPTOR ADRM1, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-18 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UFO
 
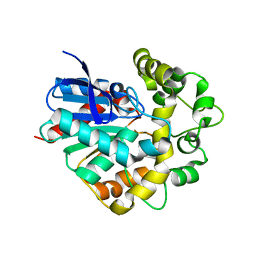 | | Laboratory evolved variant R-C1B1D33E6 of potato epoxide hydrolase StEH1 | | Descriptor: | EPOXIDE HYDROLASE | | Authors: | Carlsson, A.J, Bauer, P, Nilsson, M, Dobritzsch, D, Kamerlin, S.C.L, Widersten, M. | | Deposit date: | 2015-03-17 | | Release date: | 2016-04-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Laboratory Evolved Enzymes Provide Snapshots of the Development of Enantioconvergence in Enzyme-Catalyzed Epoxide Hydrolysis.
Chembiochem, 17, 2016
|
|
4UGA
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-((3-(((2-(3-fluorophenyl)ethyl)amino)methyl)phenoxy)methyl)-4- methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-{[3-({[2-(3-fluorophenyl)ethyl]amino}methyl)phenoxy]methyl}-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UG5
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-(2-(5-(2-(2-amino-6-methylpyridin-4-yl)ethyl)pyridin-3-yl)ethyl)-4- methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{5-[2-(2-amino-6-methylpyridin-4-yl)ethyl]pyridin-3-yl}ethyl)-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UGI
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with N1-(6-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)pyridin-2-yl)-N1,N2- dimethylethane-1,2-diamine | | Descriptor: | CHLORIDE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UGJ
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-(methyl(2-(methylamino) ethyl)amino)benzonitrile | | Descriptor: | 3-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL)-5-(METHYL(2-(METHYLAMINO)ETHYL)AMINO)BENZONITRILE, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UC9
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal dipeptide of the Phosphoprotein | | Descriptor: | ASPARTIC ACID, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UCD
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with the Nucleoprotein Phosphoprotein interaction inhibitor M81 | | Descriptor: | 1-[(2-chlorophenyl)methyl]pyrazole-3,5-dicarboxylic acid, NUCLEOPROTEIN, SULFATE ION | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
4UE6
 
 | | Structure of methylene blue-treated anaerobically purified D. fructosovorans NiFe-hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | [Nife]-Hydrogenases Revisited: Nickel-Carboxamido Bond Formation in a Variant with Accrued O2-Tolerance and a Tentative Re-Interpretation of Ni-Si States.
Metallomics, 7, 2015
|
|
4UG3
 
 | | B. subtilis GpsB N-terminal Domain | | Descriptor: | CELL CYCLE PROTEIN GPSB | | Authors: | Rismondo, J, Cleverley, R.M, Lane, H.V, Grohennig, S, Steglich, A, Moller, L, Krishna Mannala, G, Hain, T, Lewis, R.J, Halbedel, S. | | Deposit date: | 2015-03-21 | | Release date: | 2015-11-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Bacterial Cell Division Determinant Gpsb and its Interaction with Penicillin Binding Proteins.
Mol.Microbiol., 99, 2016
|
|
4UGD
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6-((((2S)-1-amino-4-((6-amino-4-methylpyridin-2-yl)methoxy)butan-2-yl) oxy)methyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-[({(2S)-1-amino-4-[(6-amino-4-methylpyridin-2-yl)methoxy]butan-2-yl}oxy)methyl]-4-methylpyridin-2-amine, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UCN
 
 | |
4UDT
 
 | | T cell receptor (TRAV22,TRBV7-9) structure | | Descriptor: | GLYCEROL, PROTEIN TRBV7-9, T-CELL RECEPTOR BETA-2 CHAIN C REGION, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2014-12-11 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of Staphylococcal Enterotoxin E in Complex with Tcr Defines the Role of Tcr Loop Positioning in Superantigen Recognition.
Plos One, 10, 2015
|
|
4UEW
 
 | | Structure of H2-treated anaerobically purified D. fructosovorans NiFe- hydrogenase | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, GLYCEROL, ... | | Authors: | Volbeda, A, Martin, L, Liebgott, P.-P, Fontecilla-Camps, J.C. | | Deposit date: | 2014-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | [NiFe]-hydrogenases revisited: nickel-carboxamido bond formation in a variant with accrued O2-tolerance and a tentative re-interpretation of Ni-SI states.
Metallomics, 7, 2015
|
|
4UF5
 
 | | Crystal structure of UCH-L5 in complex with inhibitory fragment of INO80G | | Descriptor: | NUCLEAR FACTOR RELATED TO KAPPA-B-BINDING PROTEIN, UBIQUITIN CARBOXYL-TERMINAL HYDROLASE ISOZYME L5 | | Authors: | Sahtoe, D.D, Van Dijk, W.J, El Oualid, F, Ekkebus, R, Ovaa, H, Sixma, T.K. | | Deposit date: | 2014-12-23 | | Release date: | 2015-03-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Mechanism of Uch-L5 Activation and Inhibition by Deubad Domains in Rpn13 and Ino80G.
Mol.Cell, 57, 2015
|
|
4UGE
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 4-methyl-6-((3-(piperidin-4-ylmethoxy)phenoxy)methyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-{[3-(piperidin-4-ylmethoxy)phenoxy]methyl}pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UGC
 
 | | Structure of Bacillus subtilis Nitric Oxide Synthase in complex with 6,6'-(((2S)-3-aminopropane-1,2-diyl)bis(oxymethanediyl))bis(4- methylpyridin-2-amine) | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6,6'-{[(2S)-3-aminopropane-1,2-diyl]bis(oxymethanediyl)}bis(4-methylpyridin-2-amine), CHLORIDE ION, ... | | Authors: | Holden, J.K, Poulos, T.L. | | Deposit date: | 2015-03-22 | | Release date: | 2015-06-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Inhibitor Bound Crystal Structures of Bacterial Nitric Oxide Synthase.
Biochemistry, 54, 2015
|
|
4UC8
 
 | | N-terminal globular domain of the RSV Nucleoprotein in complex with C- terminal phenylalanine of the Phosphoprotein | | Descriptor: | GLYCEROL, NUCLEOPROTEIN, PHENYLALANINE, ... | | Authors: | Ouizougun-Oubari, M, Pereira, N, Tarus, B, Galloux, M, Tortorici, M.-A, Hoos, S, Baron, B, England, P, Bontems, F, Rey, F.A, Eleouet, J.-F, Sizun, C, Slama-Schwok, A, Duquerroy, S. | | Deposit date: | 2014-12-03 | | Release date: | 2015-08-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Druggable Pocket at the Nucleocapsid/Phosphoprotein Interaction Site of the Human Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
