6X3F
 
 | | hEAAT3-IFS-Apo | | Descriptor: | CHOLINE ION, Excitatory amino acid transporter 3 | | Authors: | Qiu, B, Matthies, D, Boudker, O. | | Deposit date: | 2020-05-21 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo-EM structures of excitatory amino acid transporter 3 visualize coupled substrate, sodium, and proton binding and transport.
Sci Adv, 7, 2021
|
|
8BX7
 
 | | Structure of the rod CNG channel bound to calmodulin | | Descriptor: | Cyclic nucleotide-gated cation channel beta-1, cGMP-gated cation channel alpha-1, calmodulin-1 | | Authors: | Marino, J. | | Deposit date: | 2022-12-08 | | Release date: | 2023-04-12 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Structural basis of calmodulin modulation of the rod cyclic nucleotide-gated channel.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6WKT
 
 | |
3U58
 
 | | Crystal Structure of the Tetrahymena telomerase processivity factor Teb1 AB | | Descriptor: | DNA (5'-D(*GP*GP*GP*T)-3'), Tetrahymena Teb1 AB | | Authors: | Zeng, Z, Huang, J, Yang, Y, Lei, M. | | Deposit date: | 2011-10-11 | | Release date: | 2011-12-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.613 Å) | | Cite: | Structural basis for Tetrahymena telomerase processivity factor Teb1 binding to single-stranded telomeric-repeat DNA.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
8GC3
 
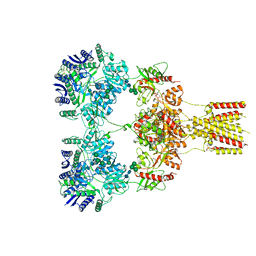 | | Domote-bound GluK2 kainate receptors in partially-open conformation 2 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8BSC
 
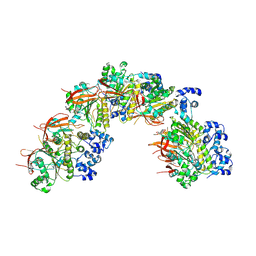 | |
8GC5
 
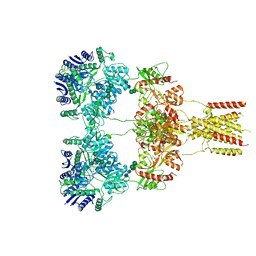 | | Domoate-bound GluK2 kainate receptors in non-active conformation | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8BQ2
 
 | | CryoEM structure of the pre-synaptic RAD51 nucleoprotein filament in the presence of ATP and Ca2+ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (30-MER), ... | | Authors: | Appleby, R, Bollschweiler, D, Pellegrini, L. | | Deposit date: | 2022-11-18 | | Release date: | 2023-05-03 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A metal ion-dependent mechanism of RAD51 nucleoprotein filament disassembly.
Iscience, 26, 2023
|
|
8GC2
 
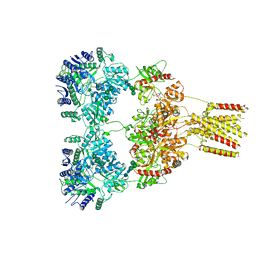 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 1 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
8GC4
 
 | | Domoate-bound GluK2 kainate receptor in partially-open conformation 3 | | Descriptor: | (2S,3S,4S)-2-CARBOXY-4-[(1Z,3E,5R)-5-CARBOXY-1-METHYL-1,3-HEXADIENYL]-3-PYRROLIDINEACETIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bogdanovic, N, Tajima, N. | | Deposit date: | 2023-03-01 | | Release date: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Structural basis for kainate receptor activation by a partial agonist
To Be Published
|
|
7QZO
 
 | | Crystal structure of GacS D1 domain | | Descriptor: | CADMIUM ION, GLYCEROL, Histidine kinase | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-31 | | Release date: | 2022-07-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
5LHU
 
 | | ATP Phosphoribosyltransferase from Mycobacterium tuberculosis in complex with the allosteric inhibitor L-Histidine | | Descriptor: | ATP phosphoribosyltransferase, GLYCEROL, HISTIDINE, ... | | Authors: | de Chiara, C, Pisco, J.P, de Carvalho, L.P, Smerdon, S.J, Walker, P.A, Ogrodowicz, R. | | Deposit date: | 2016-07-12 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Uncoupling conformational states from activity in an allosteric enzyme.
Nat Commun, 8, 2017
|
|
7QZ2
 
 | | Crystal structure of GacS D1 domain in complex with BeF3- | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, CADMIUM ION, Histidine kinase, ... | | Authors: | Fadel, F, Bassim, V, Botzanowski, T, Francis, V.I, Legrand, P, Porter, S.L, Bourne, Y, Cianferani, S, Vincent, F. | | Deposit date: | 2022-01-30 | | Release date: | 2022-07-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Insights into the atypical autokinase activity of the Pseudomonas aeruginosa GacS histidine kinase and its interaction with RetS.
Structure, 30, 2022
|
|
5LRC
 
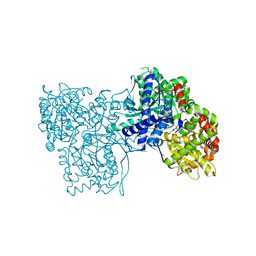 | | Crystal structure of Glycogen Phosphorylase in complex with KS114 | | Descriptor: | (1S)-1,5-anhydro-1-(5-phenyl-4H-1,2,4-triazol-3-yl)-D-glucitol, Glycogen phosphorylase, muscle form, ... | | Authors: | Kantsadi, A.L, Stravodimos, G.A, Chatzileontiadou, D.S.M, Leonidas, D.D. | | Deposit date: | 2016-08-18 | | Release date: | 2017-06-14 | | Last modified: | 2020-08-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | van der Waals interactions govern C-beta-d-glucopyranosyl triazoles' nM inhibitory potency in human liver glycogen phosphorylase.
J. Struct. Biol., 199, 2017
|
|
8BMS
 
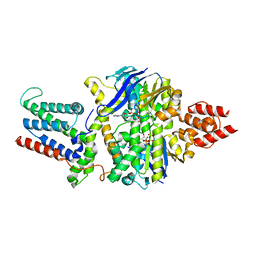 | | Cryo-EM structure of the mutant solitary ECF module 2EQ in MSP2N2 lipid nanodiscs in the ATPase closed and ATP-bound conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, Energy-coupling factor transporter ATP-binding protein EcfA2, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
8BMP
 
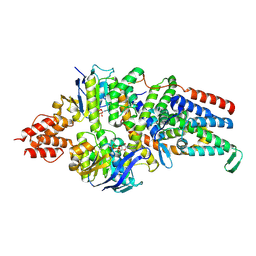 | | Cryo-EM structure of the folate-specific ECF transporter complex in MSP2N2 lipid nanodiscs bound to ATP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Energy-coupling factor transporter ATP-binding protein EcfA1, ... | | Authors: | Thangaratnarajah, C, Rheinberger, J, Paulino, C, Slotboom, D.J. | | Deposit date: | 2022-11-10 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Expulsion mechanism of the substrate-translocating subunit in ECF transporters.
Nat Commun, 14, 2023
|
|
3UF1
 
 | | Crystal Structure of Vimentin (fragment 144-251) from Homo sapiens, Northeast Structural Genomics Consortium Target HR4796B | | Descriptor: | Vimentin | | Authors: | Kuzin, A, Abashidze, M, Vorobiev, S.M, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-31 | | Release date: | 2011-11-30 | | Last modified: | 2013-04-24 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | The structure of vimentin linker 1 and rod 1B domains characterized by site-directed spin-labeling electron paramagnetic resonance (SDSL-EPR) and X-ray crystallography.
J.Biol.Chem., 287, 2012
|
|
6XGR
 
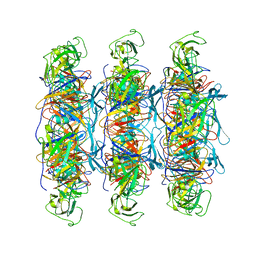 | | YSD1 major tail protein | | Descriptor: | YSD1_22 major tail protein | | Authors: | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
8BJF
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (inward-facing conformation) | | Descriptor: | ATP-binding cassette sub-family C member 4, CHOLESTEROL, CHOLESTEROL HEMISUCCINATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-11-04 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWQ
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with topotecan) | | Descriptor: | (S)-10-[(DIMETHYLAMINO)METHYL]-4-ETHYL-4,9-DIHYDROXY-1H-PYRANO[3',4':6,7]INOLIZINO[1,2-B]-QUINOLINE-3,14(4H,12H)-DIONE, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWP
 
 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with methotrexate) | | Descriptor: | ATP-binding cassette sub-family C member 4, METHOTREXATE | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWO
 
 | | Cryo-EM structure of nanodisc-reconstituted human MRP4 with E1202Q mutation (outward-facing occluded conformation) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 4, MAGNESIUM ION | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BWR
 
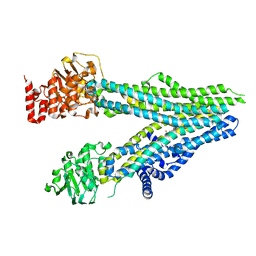 | | Cryo-EM structure of nanodisc-reconstituted wildtype human MRP4 (in complex with prostaglandin E2) | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, ATP-binding cassette sub-family C member 4 | | Authors: | Raj, I, Bloch, M, Pape, T.H, Taylor, N.M.I. | | Deposit date: | 2022-12-07 | | Release date: | 2023-08-30 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural and mechanistic basis of substrate transport by the multidrug transporter MRP4.
Structure, 31, 2023
|
|
8BGW
 
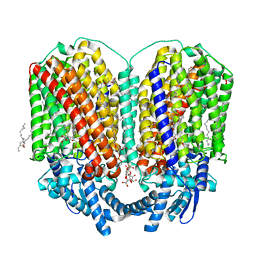 | | CryoEM structure of quinol-dependent Nitric Oxide Reductase (qNOR) from Alcaligenes xylosoxidans at 2.2 A resolution | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, CALCIUM ION, FE (III) ION, ... | | Authors: | Flynn, A, Antonyuk, S.V, Eady, R.R, Muench, S.P, Hasnain, S.S. | | Deposit date: | 2022-10-28 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | A 2.2 angstrom cryoEM structure of a quinol-dependent NO Reductase shows close similarity to respiratory oxidases.
Nat Commun, 14, 2023
|
|
6XBM
 
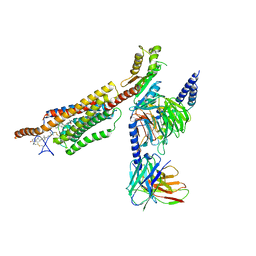 | | Structure of human SMO-Gi complex with 24(S),25-EC | | Descriptor: | 17-[3-(3,3-DIMETHYL-OXIRANYL)-1-METHYL-PROPYL]-10,13-DIMETHYL-2,3,4,7,8,9,10,11,12,13,14,15,16,17-TETRADECAHYDRO-1H-CYC LOPENTA[A]PHENANTHREN-3-OL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Qi, X, Long, T, Li, X. | | Deposit date: | 2020-06-06 | | Release date: | 2020-09-30 | | Last modified: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Sterols in an intramolecular channel of Smoothened mediate Hedgehog signaling.
Nat.Chem.Biol., 16, 2020
|
|
