6H5G
 
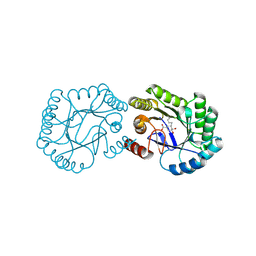 | | Crystal structure of DHQ1 from Salmonella typhi covalently modified by ligand 3 | | Descriptor: | (1~{R},3~{S},4~{R},5~{R})-3-methyl-4,5-bis(hydroxyl)cyclohexane-1-carboxylic acid, 3-dehydroquinate dehydratase | | Authors: | Sanz-Gaitero, M, Maneiro, M, Lence, E, Otero, J.M, Thompson, P, Hawkins, A.R, Gonzalez-Bello, C, van Raaij, M.J. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Hydroxylammonium Derivatives for Selective Active-site Lysine Modification in the Anti-virulence Bacterial Target DHQ1 Enzyme.
Org Chem Front, 6, 2019
|
|
6NWA
 
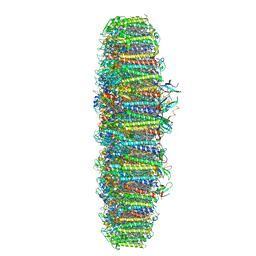 | | The structure of the photosystem I IsiA super-complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Toporik, H, Li, J, Williams, D, Chiu, P.L, Mazor, Y. | | Deposit date: | 2019-02-06 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | The structure of the stress-induced photosystem I-IsiA antenna supercomplex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6HEE
 
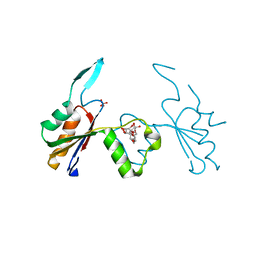 | | Crystal structure of Extracellular Domain 1 (ECD1) of FtsX from S. pneumonie in complex with undecyl-maltoside | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cell division protein FtsX, SULFATE ION, ... | | Authors: | Martinez-Caballero, S, Alcorlo-Pages, M, Hermoso, J.A. | | Deposit date: | 2018-08-20 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Large Extracellular Loop of FtsX and Its Interaction with the Essential Peptidoglycan Hydrolase PcsB in Streptococcus pneumoniae.
Mbio, 10, 2019
|
|
5MPT
 
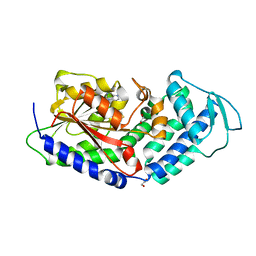 | | Structure of the citrinin polyketide synthase CMeT domain | | Descriptor: | 1,2-ETHANEDIOL, Citrinin polyketide synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Herbst, D.A, Storm, P.A, Townsend, C.A, Maier, T. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Functional and Structural Analysis of Programmed C-Methylation in the Biosynthesis of the Fungal Polyketide Citrinin.
Cell Chem Biol, 24, 2017
|
|
8D52
 
 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing (2-naphthyl)-beta-3-homoalanine | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36], Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Kreitler, D.F, Gellman, S.H. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Harnessing Aromatic-Histidine Interactions through Synergistic Backbone Extension and Side Chain Modification.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6MYO
 
 | | Avian mitochondrial complex II with flutolanyl bound | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, DI(HYDROXYETHYL)ETHER, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Berry, E.A, Huang, L.-S. | | Deposit date: | 2018-11-02 | | Release date: | 2019-11-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic investigation of the ubiquinone binding site of respiratory Complex II and its inhibitors.
Biochim Biophys Acta Proteins Proteom, 1869, 2021
|
|
8SG5
 
 | | Cytochrome P450 (CYP) 3A5 crystallized with clotrimazole | | Descriptor: | 1-[(2-CHLOROPHENYL)(DIPHENYL)METHYL]-1H-IMIDAZOLE, Cytochrome P450 3A5, DODECYL NONA ETHYLENE GLYCOL ETHER, ... | | Authors: | Hsu, M.H, Johnson, E.F. | | Deposit date: | 2023-04-11 | | Release date: | 2023-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Differential Effects of Clotrimazole on X-Ray Crystal Structures of Human Cytochromes P450 3A5 and 3A4.
Drug Metab.Dispos., 51, 2023
|
|
8VLM
 
 | | Crystal structure of the yeast cytosine deaminase (yCD) E64V-M100W heterodimer | | Descriptor: | 1,2-ETHANEDIOL, Cytosine deaminase, ZINC ION | | Authors: | Picard, M.-E, Grenier, G, Despres, P.C, Dube, A.K, Landry, C.R, Shi, R. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-21 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Compensatory mutations potentiate constructive neutral evolution by gene duplication.
Science, 385, 2024
|
|
6I6V
 
 | | SEPIAPTERIN REDUCTASE IN COMPLEX WITH COMPOUND 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-[[(3~{R})-oxan-3-yl]methylsulfonyl]-2-azaspiro[4.5]decane, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Alen, J, Schade, M, Wagener, M, Blaesse, M. | | Deposit date: | 2018-11-15 | | Release date: | 2019-07-10 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Fragment-Based Discovery of Novel Potent Sepiapterin Reductase Inhibitors.
J.Med.Chem., 62, 2019
|
|
8VHU
 
 | | Crystal structure of dATP bound E. coli class Ia ribonucleotide reductase alpha construct fused with the C-terminal tail of E. coli class Ia beta subunit | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CHLORIDE ION, Fusion protein of Ribonucleoside-diphosphate reductase 1 subunits alpha and beta, ... | | Authors: | Funk, M.A, Zimanyi, C.M, Drennan, C.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-09-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | How ATP and dATP Act as Molecular Switches to Regulate Enzymatic Activity in the Prototypical Bacterial Class Ia Ribonucleotide Reductase.
Biochemistry, 63, 2024
|
|
8OOQ
 
 | |
6R4D
 
 | | Aurora-A in complex with shape-diverse fragment 58 | | Descriptor: | (1~{S},10~{S})-12-cyclobutyl-5-methyl-1-oxidanyl-10-propan-2-yl-9,12-diazatricyclo[8.2.1.0^{2,7}]trideca-2(7),3,5-trien-11-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.009 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
5N2B
 
 | |
8RNV
 
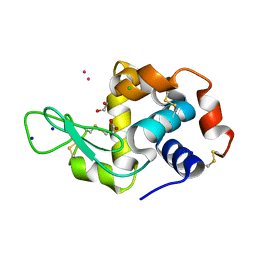 | | Hen Egg White Lysozyme soaked with cis-Ru(DMSO)4Cl2 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Oszajca, M, Flejszar, M, Szura, A, Drozdz, P, Brindell, M, Kurpiewska, K. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Exploring the coordination chemistry of ruthenium complexes with lysozymes: structural and in-solution studies.
Front Chem, 12, 2024
|
|
6YTT
 
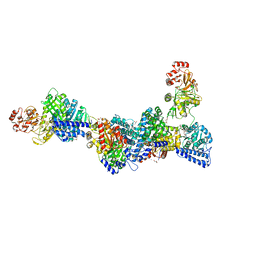 | | CO-dehydrogenase/Acetyl-CoA synthase (CODH/ACS) from Clostridium autoethanogenum at 3.0-A resolution | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CO dehydrogenase/acetyl-CoA synthase complex, beta subunit, ... | | Authors: | Wagner, T, Lemaire, O.N. | | Deposit date: | 2020-04-24 | | Release date: | 2020-11-04 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Gas channel rerouting in a primordial enzyme: Structural insights of the carbon-monoxide dehydrogenase/acetyl-CoA synthase complex from the acetogen Clostridium autoethanogenum.
Biochim Biophys Acta Bioenerg, 1862, 2020
|
|
4XPI
 
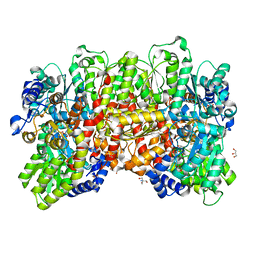 | | Fe protein independent substrate reduction by nitrogenase variants altered in intramolecular electron transfer | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-HYDROXY-3-CARBOXY-ADIPIC ACID, CALCIUM ION, ... | | Authors: | Danyal, K, Rasmusen, A.J, Keable, S.M, Shaw, S, Zadvornyy, O, Duval, S, Dean, D.R, Raugei, S, Peters, J.W, Seefeldt, L.C. | | Deposit date: | 2015-01-17 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Fe protein-independent substrate reduction by nitrogenase MoFe protein variants.
Biochemistry, 54, 2015
|
|
8TVJ
 
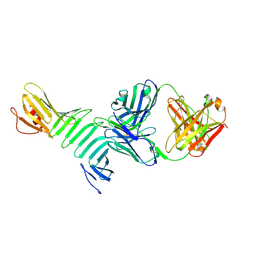 | | Fab 227-1 in complex with OspA | | Descriptor: | Heavy chain of Fab 227-1, Light chain of Fab 227-1, Outer surface protein A | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-08-18 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.168 Å) | | Cite: | Fab 227-1 in complex with OspA
To Be Published
|
|
5Y5F
 
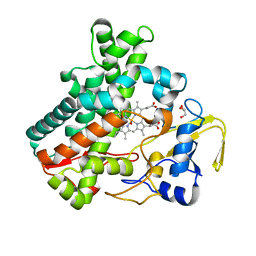 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
6R49
 
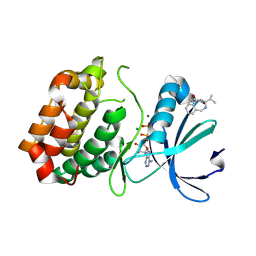 | | Aurora-A in complex with shape-diverse fragment 39 | | Descriptor: | (1~{S},10~{S})-12-cyclopropyl-1-oxidanyl-10-propan-2-yl-9,12-diazatricyclo[8.2.1.0^{2,7}]trideca-2(7),3,5-trien-11-one, ADENOSINE-5'-DIPHOSPHATE, Aurora kinase A, ... | | Authors: | Bayliss, R, McIntyre, P.J. | | Deposit date: | 2019-03-22 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.209 Å) | | Cite: | Construction of a Shape-Diverse Fragment Set: Design, Synthesis and Screen against Aurora-A Kinase.
Chemistry, 25, 2019
|
|
6OJU
 
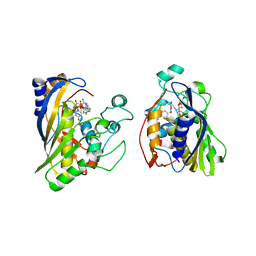 | | Crystal structure of human thymidylate synthase Delta (7-29) in complex with dUMP and 2-amino-4-oxo-4,7-dihydro-pyrrolo[2,3-d]pyrimidine-methyl-phenyl-D-glutamic acid | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, N-{4-[(2-amino-4-oxo-4,7-dihydro-3H-pyrrolo[2,3-d]pyrimidin-5-yl)methyl]benzene-1-carbonyl}-D-glutamic acid, Thymidylate synthase,Thymidylate synthase | | Authors: | Czyzyk, D.J, Anderson, K.S, Valhondo, M, Jorgensen, W.L. | | Deposit date: | 2019-04-12 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.884 Å) | | Cite: | Understanding the structural basis of species selective, stereospecific inhibition for Cryptosporidium and human thymidylate synthase.
Febs Lett., 593, 2019
|
|
6RC0
 
 | |
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
8PE1
 
 | | Crystal structure of Gel4 in complex with Nanobody 4 | | Descriptor: | 1,3-beta-glucanosyltransferase, 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody 4, ... | | Authors: | Macias-Leon, J, Redrado-Hernandez, S, Castro-Lopez, J, Sanz, A.B, Arias, M, Farkas, V, Vincke, C, Muyldermans, S, Pardo, J, Arroyo, J, Galvez, E, Hurtado-Guerrero, R. | | Deposit date: | 2023-06-13 | | Release date: | 2024-06-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Broad Protection against Invasive Fungal Disease from a Nanobody Targeting the Active Site of Fungal beta-1,3-Glucanosyltransferases.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8SFM
 
 | | Crystal structure of the engineered SsoPox variant IVB10 in alternate state | | Descriptor: | 1,2-ETHANEDIOL, Aryldialkylphosphatase, COBALT (II) ION, ... | | Authors: | Jacquet, P, Billot, R, Shimon, A, Hoekstra, N, Bergonzi, C, Jenks, A, Daude, D, Elias, M.H. | | Deposit date: | 2023-04-11 | | Release date: | 2024-04-17 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Changes in Active Site Loop Conformation Relate to the Transition toward a Novel Enzymatic Activity.
Jacs Au, 4, 2024
|
|
8UL9
 
 | | Cholinephosphotransferase in complex with diacylglycerol | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, Cholinephosphotransferase 1, DIACYL GLYCEROL, ... | | Authors: | Roberts, J.R, Maeda, S, Ohi, M.D. | | Deposit date: | 2023-10-16 | | Release date: | 2024-10-23 | | Last modified: | 2025-01-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for catalysis and selectivity of phospholipid synthesis by eukaryotic choline-phosphotransferase.
Nat Commun, 16, 2025
|
|
