6RQD
 
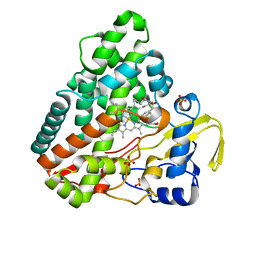 | | CYP121 in complex with 3-chloro dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(3-chloranyl-4-oxidanyl-phenyl)methyl]-6-[(4-hydroxyphenyl)methyl]piperazine-2,5-dione, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.499 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6XTA
 
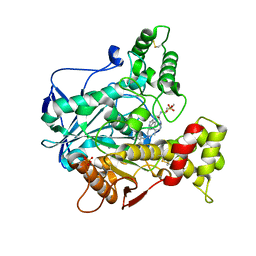 | | Recombinant human butyrylcholinesterase in complex with (2S)-N-[2-(1-benzylazepan-4-yl)ethyl]-2-(butylamino)-3-(1H-indol-3-yl)propanamide | | Descriptor: | (2~{S})-2-(butylamino)-3-(1~{H}-indol-3-yl)-~{N}-[2-[(4~{R})-1-(phenylmethyl)azepan-4-yl]ethyl]propanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Nachon, F, Meden, A, Knez, D, Gobec, S. | | Deposit date: | 2020-01-15 | | Release date: | 2020-10-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-activity relationship study of tryptophan-based butyrylcholinesterase inhibitors.
Eur.J.Med.Chem., 208, 2020
|
|
4ZVR
 
 | |
7PC4
 
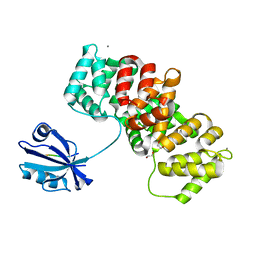 | | The PDZ domain of SNTB1 complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | 1,2-ETHANEDIOL, Beta-1-syntrophin,Annexin A2, CALCIUM ION, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC5
 
 | | The third PDZ domain of PDZD7 complexed with the PDZ-binding motif of EXOC4 | | Descriptor: | CALCIUM ION, Exocyst complex component 4, GLYCEROL, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PCB
 
 | | The PDZ domain of SNX27 fused with ANXA2 | | Descriptor: | CALCIUM ION, GLYCEROL, Sorting nexin-27,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC8
 
 | | The PDZ domain of SNTG1 complexed with the phosphomimetic mutant PDZ-binding motif of RSK1 | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC7
 
 | | The PDZ domain of SNTG1 complexed with the acetylated PDZ-binding motif of PTEN | | Descriptor: | CALCIUM ION, GLYCEROL, Gamma-1-syntrophin,Annexin A2, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7PC9
 
 | | The PDZ domain of SYNJ2BP complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Protein Tax-1, Synaptojanin-2-binding protein,Annexin A2 | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6XSV
 
 | |
7PC3
 
 | | The second PDZ domain of DLG1 complexed with the PDZ-binding motif of HTLV1-TAX1 | | Descriptor: | CALCIUM ION, Disks large homolog 1,Annexin A2, GLYCEROL, ... | | Authors: | Cousido-Siah, A, Trave, G, Gogl, G. | | Deposit date: | 2021-08-03 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A scalable strategy to solve structures of PDZ domains and their complexes.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6XYR
 
 | | Structure of the T4Lnano fusion protein | | Descriptor: | CALCIUM ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Benoit, R.M, Bierig, T, Collu, C, Engilberge, S, Olieric, V. | | Deposit date: | 2020-01-31 | | Release date: | 2020-12-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Chimeric single alpha-helical domains as rigid fusion protein connections for protein nanotechnology and structural biology.
Structure, 30, 2022
|
|
4ZVT
 
 | |
6XXU
 
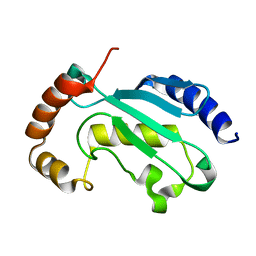 | | Solution NMR structure of the native form of UbcH7 (UBE2L3) | | Descriptor: | Ubiquitin-conjugating enzyme E2 L3 | | Authors: | Marousis, K.D, Seliami, A, Birkou, M, Episkopou, V, Spyroulias, G.A. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H,13C,15N backbone and side-chain resonance assignment of the native form of UbcH7 (UBE2L3) through solution NMR spectroscopy.
Biomol.Nmr Assign., 14, 2020
|
|
6XXW
 
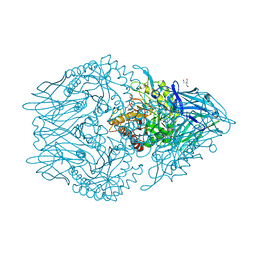 | | Structure of beta-D-Glucuronidase for Dictyoglomus thermophilum. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucuronidase, ... | | Authors: | Lafite, P, Daniellou, R. | | Deposit date: | 2020-01-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Thioglycoligation of aromatic thiols using a natural glucuronide donor.
Org.Biomol.Chem., 18, 2020
|
|
6RQ1
 
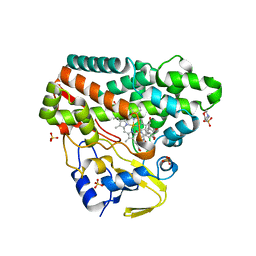 | | CYP121 in complex with 2-methyl dicyclotyrosine | | Descriptor: | (3~{S},6~{S})-3-[(4-hydroxyphenyl)methyl]-6-[(2-methyl-4-oxidanyl-phenyl)methyl]piperazine-2,5-dione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Mycocyclosin synthase, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
6RQB
 
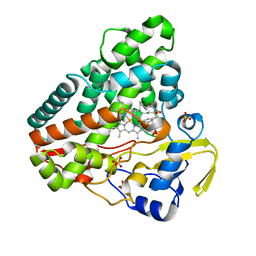 | | CYP121 in complex with 3-bromo dicyclotyrosine | | Descriptor: | 3-bromo dicyclotyrosine, Mycocyclosin synthase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Poddar, H, Levy, C. | | Deposit date: | 2019-05-15 | | Release date: | 2020-04-22 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.459 Å) | | Cite: | Structure-Activity Relationships of cyclo (l-Tyrosyl-l-tyrosine) Derivatives Binding to Mycobacterium tuberculosis CYP121: Iodinated Analogues Promote Shift to High-Spin Adduct.
J.Med.Chem., 62, 2019
|
|
3HM7
 
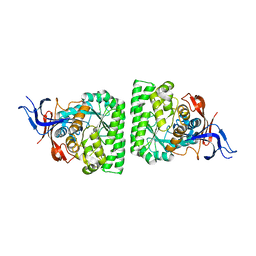 | | Crystal structure of allantoinase from Bacillus halodurans C-125 | | Descriptor: | Allantoinase, ZINC ION | | Authors: | Patskovsky, Y, Romero, R, Rutter, M, Miller, S, Wasserman, S.R, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York Structural GenomiX Research Consortium (NYSGXRC), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Allantoinase from Bacillus Halodurans
To be Published
|
|
6XXY
 
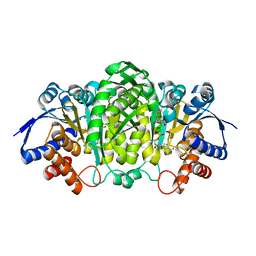 | | Crystal structure of Haemophilus influenzae 3-isopropylmalate dehydrogenase in complex with O-isobutenyl oxalylhydroxamate. | | Descriptor: | 2-(2-methylprop-2-enoxyamino)-2-oxidanylidene-ethanoic acid, 3-isopropylmalate dehydrogenase, MAGNESIUM ION, ... | | Authors: | Miggiano, R, Rossi, F, Martignon, S, Rizzi, M. | | Deposit date: | 2020-01-28 | | Release date: | 2020-02-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of Haemophilus influenzae 3-isopropylmalate dehydrogenase (LeuB) in complex with the inhibitor O-isobutenyl oxalylhydroxamate.
Biochem.Biophys.Res.Commun., 524, 2020
|
|
6XOF
 
 | | Crystal structure of SCLam, a non-specific endo-beta-1,3(4)-glucanase from family GH16 | | Descriptor: | CALCIUM ION, GH16 family protein, GLYCEROL | | Authors: | Liberato, M.V, Bernardes, A, Polikarpov, I, Squina, F. | | Deposit date: | 2020-07-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQH
 
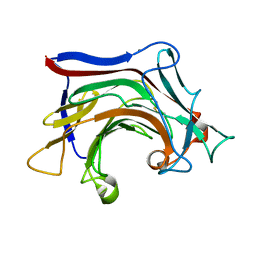 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with cellotriose, presenting a 1,3-beta-D-cellobiosyl-glucose and a cellobiose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQM
 
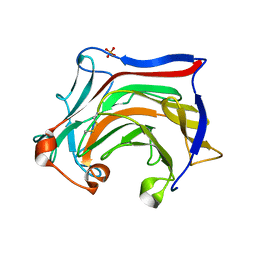 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with laminarihexaose, presenting a laminaribiose and a glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, PHOSPHATE ION, ... | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XQG
 
 | | Crystal structure of SCLam E144S mutant, a non-specific endo-beta-1,3(4)-glucanase from family GH16, co-crystallized with 1,3-beta-D-cellobiosyl-cellobiose, presenting a 1,3-beta-D-cellobiosyl-glucose at active site | | Descriptor: | CALCIUM ION, GH16 family protein, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose | | Authors: | Liberato, M.V, Squina, F. | | Deposit date: | 2020-07-09 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the dual cleavage activity of the GH16 laminarinase enzyme class on beta-1,3 and beta-1,4 glycosidic bonds.
J.Biol.Chem., 296, 2021
|
|
6XDS
 
 | |
5XV6
 
 | | Crystal structure of ATG101-ATG13HORMA | | Descriptor: | Autophagy-related protein 101, Autophagy-related protein 13 | | Authors: | Kim, B.-W, Song, H.K. | | Deposit date: | 2017-06-26 | | Release date: | 2018-07-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | The C-terminal region of ATG101 bridges ULK1 and PtdIns3K complex in autophagy initiation.
Autophagy, 14, 2018
|
|
