3NNR
 
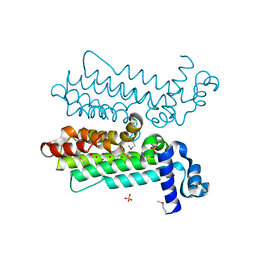 | |
3OB8
 
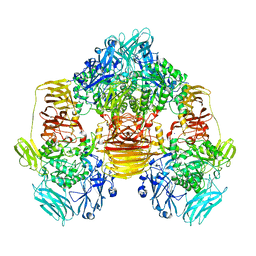 | | Structure of the beta-galactosidase from Kluyveromyces lactis in complex with galactose | | Descriptor: | Beta-galactosidase, MAGNESIUM ION, MANGANESE (II) ION, ... | | Authors: | Fernandez-Leiro, R, Pereira-Rodriguez, A, Becerra, M, Gonzalez-Siso, I, Cerdan, M.E, Sanz-Aparicio, J. | | Deposit date: | 2010-08-06 | | Release date: | 2011-08-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of specificity in tetrameric Kluyveromyces lactis beta-galactosidase.
J.Struct.Biol., 177, 2012
|
|
3OCZ
 
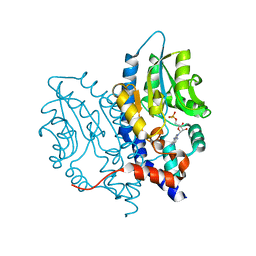 | | Structure of Recombinant Haemophilus influenzae e(P4) Acid Phosphatase Complexed with the inhibitor adenosine 5-O-thiomonophosphate | | Descriptor: | ADENOSINE -5'-THIO-MONOPHOSPHATE, Lipoprotein E, MAGNESIUM ION | | Authors: | Singh, H, Schuermann, J, Reilly, T, Calcutt, M, Tanner, J. | | Deposit date: | 2010-08-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of the inhibition of class C acid phosphatases by adenosine 5'-phosphorothioate.
Febs J., 278, 2011
|
|
3QTH
 
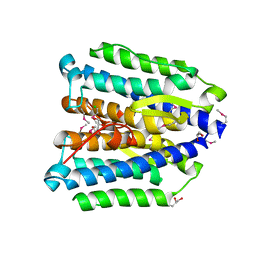 | |
3QVK
 
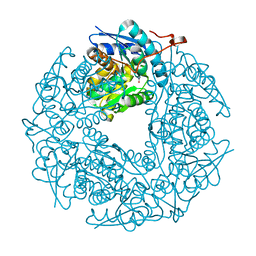 | | Allantoin racemase from Klebsiella pneumoniae | | Descriptor: | 1-(2,5-DIOXO-2,5-DIHYDRO-1H-IMIDAZOL-4-YL)UREA, Putative hydantoin racemase | | Authors: | French, J.B, Neau, D.B, Ealick, S.E. | | Deposit date: | 2011-02-25 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.999 Å) | | Cite: | Characterization of the Structure and Function of Klebsiella pneumoniae Allantoin Racemase.
J.Mol.Biol., 410, 2011
|
|
3QW4
 
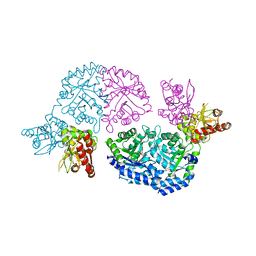 | | Structure of Leishmania donovani UMP synthase | | Descriptor: | CHLORIDE ION, UMP synthase, URIDINE-5'-MONOPHOSPHATE | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
3QW3
 
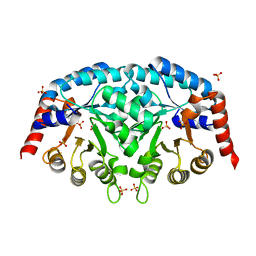 | | Structure of Leishmania donovani OMP decarboxylase | | Descriptor: | Orotidine-5-phosphate decarboxylase/orotate phosphoribosyltransferase, putative (Ompdcase-oprtase, putative), ... | | Authors: | French, J.B, Ealick, S.E. | | Deposit date: | 2011-02-26 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Leishmania donovani UMP synthase is essential for promastigote viability and has an unusual tetrameric structure that exhibits substrate-controlled oligomerization.
J.Biol.Chem., 286, 2011
|
|
3QWR
 
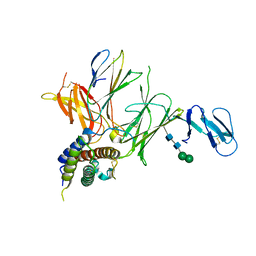 | | Crystal structure of IL-23 in complex with an adnectin | | Descriptor: | ADNECTIN, Interleukin-12 subunit beta, Interleukin-23 subunit alpha, ... | | Authors: | Wei, A, Sheriff, S. | | Deposit date: | 2011-02-28 | | Release date: | 2012-02-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structures of adnectin/protein complexes reveal an expanded binding footprint.
Structure, 20, 2012
|
|
3QYK
 
 | |
3QXD
 
 | | F54C HLA-DR1 bound with CLIP peptide | | Descriptor: | HLA class II histocompatibility antigen gamma chain peptide, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Painter, C.A, Stern, L.J. | | Deposit date: | 2011-03-01 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QZL
 
 | |
3QVY
 
 | | Crystal structure of the Zn-RIDC1 complex stabilized by BMOE crosslinks | | Descriptor: | 1,1'-ethane-1,2-diylbis(1H-pyrrole-2,5-dione), Cytochrome cb562, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Salgado, E.N, Tezcan, F.A. | | Deposit date: | 2011-02-26 | | Release date: | 2011-06-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Templated construction of a zn-selective protein dimerization motif.
Inorg.Chem., 50, 2011
|
|
3QXA
 
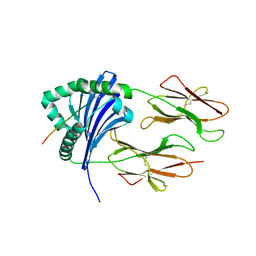 | | HLA-DR1 bound with CLIP peptide | | Descriptor: | HLA class II histocompatibility antigen gamma chain peptide, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | Painter, C.A, Stern, L.J. | | Deposit date: | 2011-03-01 | | Release date: | 2011-11-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | Conformational lability in the class II MHC 310 helix and adjacent extended strand dictate HLA-DM susceptibility and peptide exchange.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3QYT
 
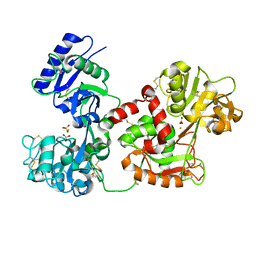 | | Diferric bound human serum transferrin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, FE (III) ION, ... | | Authors: | Yang, N, Zhang, H, Wang, M, Hao, Q, Sun, H. | | Deposit date: | 2011-03-03 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Iron and bismuth bound human serum transferrin reveals a partially-opened conformation in the N-lobe
Sci Rep, 2, 2012
|
|
3QZB
 
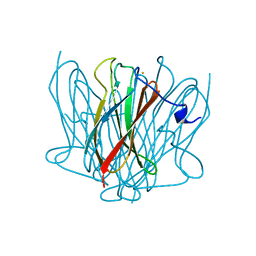 | |
3QZO
 
 | | Staphylococcus aureus IsdA NEAT domain in complex with heme, reduced crystal | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Grigg, J.C, Mao, C.X, Murphy, M.E.P. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Iron-Coordinating Tyrosine Is a Key Determinant of NEAT Domain Heme Transfer.
J.Mol.Biol., 413, 2011
|
|
3QZS
 
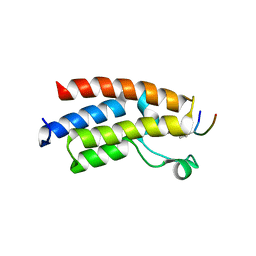 | |
3QZN
 
 | | Staphylococcus aureus IsdA NEAT domain Y166A variant in complex with heme | | Descriptor: | GLYCEROL, Iron-regulated surface determinant protein A, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Grigg, J.C, Mao, C.X, Murphy, M.E.P. | | Deposit date: | 2011-03-06 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Iron-Coordinating Tyrosine Is a Key Determinant of NEAT Domain Heme Transfer.
J.Mol.Biol., 413, 2011
|
|
3RDO
 
 | | Crystal structure of R7-2 streptavidin complexed with biotin | | Descriptor: | BIOTIN, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-01 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RE5
 
 | | Crystal structure of R4-6 streptavidin | | Descriptor: | GLYCEROL, PENTAETHYLENE GLYCOL, Streptavidin | | Authors: | Malashkevich, V.N, Magalhaes, M, Czecster, C.M, Guan, R, Levy, M, Almo, S.C. | | Deposit date: | 2011-04-02 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Evolved streptavidin mutants reveal key role of loop residue in high-affinity binding.
Protein Sci., 20, 2011
|
|
3RIN
 
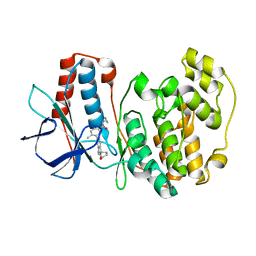 | | p38 kinase crystal structure in complex with small molecule inhibitor | | Descriptor: | Mitogen-activated protein kinase 14, N-cyclopropyl-4-methyl-3-(2'-oxo-1',2'-dihydrospiro[cyclopentane-1,3'-indol]-6'-yl)benzamide | | Authors: | Segarra, V, Eastwood, P, Roca, R, Fisher, M, Lamers, M. | | Deposit date: | 2011-04-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Indolin-2-one p38(alpha) inhibitors I: design, profiling and crystallographic binding mode.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3R09
 
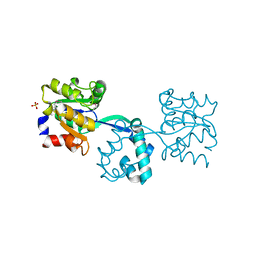 | | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg | | Descriptor: | Hydrolase, haloacid dehalogenase-like family, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Dunaway-Mariano, D, Allen, K.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI), New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2011-03-07 | | Release date: | 2011-04-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of probable HAD family hydrolase from Pseudomonas fluorescens Pf-5 with bound Mg
To be Published
|
|
3R0U
 
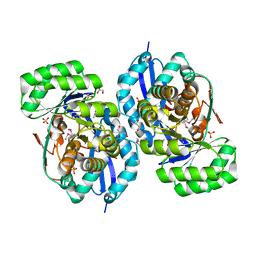 | | Crystal structure of NYSGRC enolase target 200555, a putative dipeptide epimerase from Francisella philomiragia : Tartrate and Mg complex | | Descriptor: | D(-)-TARTARIC ACID, Enzyme of enolase superfamily, GLYCEROL, ... | | Authors: | Vetting, M.W, Hillerich, B, Seidel, R.D, Zencheck, W.D, Toro, R, Imker, H.J, Gerlt, J.A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2011-03-09 | | Release date: | 2011-04-06 | | Last modified: | 2012-03-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Homology models guide discovery of diverse enzyme specificities among dipeptide epimerases in the enolase superfamily.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
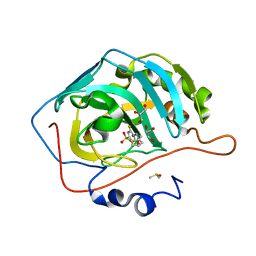
3R17
 
 | |
3RKU
 
 | |
