8JWV
 
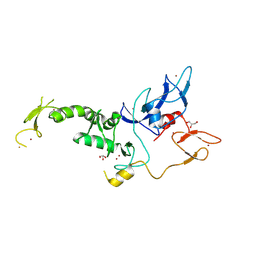 | | Untethered R0RBR | | Descriptor: | BARIUM ION, E3 ubiquitin-protein ligase parkin, GLYCEROL, ... | | Authors: | Lenka, D.R, Kumar, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Additional feedforward mechanism of Parkin activation via binding of phospho-UBL and RING0 in trans.
Elife, 13, 2024
|
|
6G1D
 
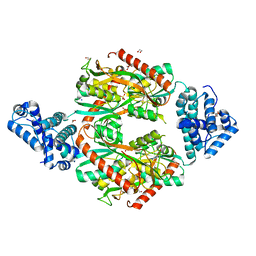 | | Corynebacterium glutamicum OxyR C206 mutant | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Young, D.R, Pedre, B.P, Messens, J.M. | | Deposit date: | 2018-03-21 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.992 Å) | | Cite: | Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
5HI4
 
 | | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists | | Descriptor: | (9'S,17'R)-6'-chloro-N-methyl-9'-{[(1-methyl-1H-pyrazol-5-yl)carbonyl]amino}-10',19'-dioxo-2'-oxa-11',18'-diazaspiro[cyclopentane-1,21'-tetracyclo[20.2.2.2~12,15~.1~3,7~]nonacosane]-1'(24'),3'(29'),4',6',12',14',22',25',27'-nonaene-17'-carboxamide, CAT-2000 FAB heavy chain, CAT-2000 FAB light chain, ... | | Authors: | Liu, S. | | Deposit date: | 2016-01-11 | | Release date: | 2016-08-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Binding site elucidation and structure guided design of macrocyclic IL-17A antagonists.
Sci Rep, 6, 2016
|
|
6PGJ
 
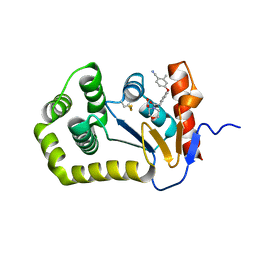 | | Crystal Structure of EcDsbA in a complex with unpurified reaction product A5 (Morpholine carboxylic acid 7) | | Descriptor: | (3R)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, (3S)-4-{[4-(4-cyano-3-methylphenoxy)phenyl]acetyl}morpholine-3-carboxylic acid, COPPER (II) ION, ... | | Authors: | Ilyichova, O.V, Bentley, M, Doak, B, Scanlon, M.J. | | Deposit date: | 2019-06-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rapid Elaboration of Fragments into Leads by X-ray Crystallographic Screening of Parallel Chemical Libraries (REFiL X ).
J.Med.Chem., 63, 2020
|
|
6G1Y
 
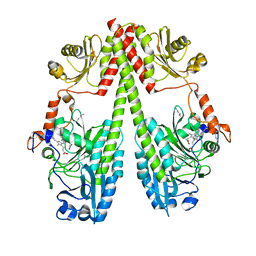 | | Crystal structure of the photosensory core module (PCM) of a bathy phytochrome from Agrobacterium fabrum in the Pfr state. | | Descriptor: | 3-[(2Z)-2-({3-(2-carboxyethyl)-5-[(E)-(4-ethenyl-3-methyl-5-oxo-1,5-dihydro-2H-pyrrol-2-ylidene)methyl]-4-methyl-1H-pyrrol-2-yl}methylidene)-5-{(Z)-[(3E,4S)-3-ethylidene-4-methyl-5-oxopyrrolidin-2-ylidene]methyl}-4-methyl-2H-pyrrol-3-yl]propanoic acid, Bacteriophytochrome protein | | Authors: | Schmidt, A, Qureshi, B.M, Scheerer, P. | | Deposit date: | 2018-03-22 | | Release date: | 2018-11-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural snapshot of a bacterial phytochrome in its functional intermediate state.
Nat Commun, 9, 2018
|
|
7CB6
 
 | |
6XK3
 
 | |
8JXK
 
 | | Crystal Structure of Rv0047c from Mycobacterium tuberculosis | | Descriptor: | Conserved protein | | Authors: | Ansari, M.S, Yadav, V, Zohib, M, Pal, R.K, Biswal, B.K, Arora, A. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structure of Rv0047c from Mycobacterium tuberculosis
To Be Published
|
|
6G5P
 
 | | Crystal structure of human SP100 in complex with bromodomain-focused fragment FM009493b 2,3-Dimethoxy-2,3-dimethyl-2,3-dihydro-1,4-benzodioxin-6-amine | | Descriptor: | (2~{R},3~{R})-2,3-dimethoxy-2,3-dimethyl-1,4-benzodioxin-6-amine, 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Talon, R.P.H, Krojer, T, Tallant, C, Nunez-Alonso, G, Fairhead, M, Szykowska, A, Collins, P, Pearce, N.M, Ng, J, MacLean, E, Wright, N, Douangamath, A, Brandao-Neto, J, Burgess-Brown, N, Huber, K, Knapp, S, Brennan, P.E, Arrowsmith, C.H, Edwards, A.M, Bountra, C, von Delft, F. | | Deposit date: | 2018-03-29 | | Release date: | 2018-04-11 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Identifying small molecule binding sites for epigenetic proteins at domain-domain interfaces
Biorxiv, 2018
|
|
5W0Z
 
 | |
7CKH
 
 | | Crystal structure of TMSiPheRS | | Descriptor: | Tyrosine--tRNA ligase | | Authors: | Sun, J.P, Wang, J.Y, Zhu, Z.L, He, Q.T, Xiao, P. | | Deposit date: | 2020-07-17 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.79492676 Å) | | Cite: | DeSiphering receptor core-induced and ligand-dependent conformational changes in arrestin via genetic encoded trimethylsilyl 1 H-NMR probe.
Nat Commun, 11, 2020
|
|
5VPW
 
 | | Nitrogenase Cp1 at pH 5 | | Descriptor: | 3-HYDROXY-3-CARBOXY-ADIPIC ACID, FE (II) ION, FE(8)-S(7) CLUSTER, ... | | Authors: | Morrison, C.N, Spatzal, T, Rees, D.C. | | Deposit date: | 2017-05-05 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reversible Protonated Resting State of the Nitrogenase Active Site.
J. Am. Chem. Soc., 139, 2017
|
|
5H61
 
 | | Structure of Transferase mutant-C23S,C199S | | Descriptor: | Transferase | | Authors: | Park, J.B, Yoo, Y, Kim, J. | | Deposit date: | 2016-11-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural basis for arginine glycosylation of host substrates by bacterial effector proteins.
Nat Commun, 9, 2018
|
|
8JV4
 
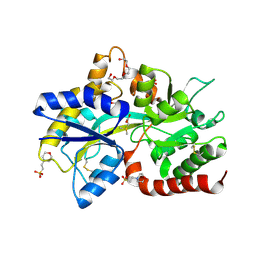 | | Structure of the SAR11 PotD in complex with DMSP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, GLYCEROL, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Structure of the SAR11 PotD in complex with DMSP
To Be Published
|
|
6UQ0
 
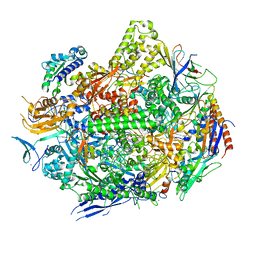 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 4 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5VQE
 
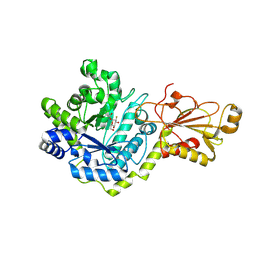 | | Beta-glucoside phosphorylase BglX bound to 2FGlc | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucoside phosphorylase BglX | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
6XJN
 
 | |
6PHZ
 
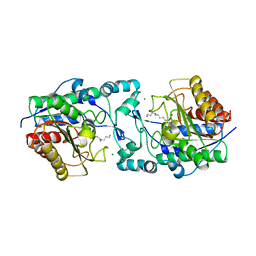 | | Crystal structure of Marinobacter subterrani acetylpolyamine amidohydrolase (msAPAH) complexed with 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptan-2-one | | Descriptor: | 7-[(3-aminopropyl)amino]-1,1,1-trifluoroheptane-2,2-diol, Acetylpolyamine Amidohydrolase, MAGNESIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2019-06-25 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of the Acetylpolyamine Amidohydrolase from the Deep Earth HalophileMarinobacter subterrani.
Biochemistry, 58, 2019
|
|
6G61
 
 | |
6UX8
 
 | |
6G65
 
 | |
6G6C
 
 | |
8JV1
 
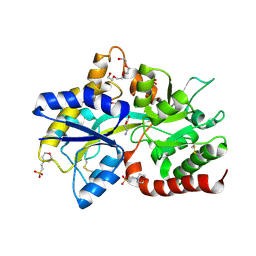 | | Structure of the SAR11 PotD in complex with GABA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Structure of the SAR11 PotD in complex with GABA
To Be Published
|
|
6G6P
 
 | | Crystal structure of the computationally designed Ika8 protein: crystal packing No.2 in P63 | | Descriptor: | Ika8 | | Authors: | Noguchi, H, Addy, C, Simoncini, D, Van Meervelt, L, Schiex, T, Zhang, K.Y.J, Tame, J.R.H, Voet, A.R.D. | | Deposit date: | 2018-04-01 | | Release date: | 2018-11-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Computational design of symmetrical eight-bladed beta-propeller proteins.
IUCrJ, 6, 2019
|
|
5VRG
 
 | |
