8UZN
 
 | |
8USK
 
 | |
8USE
 
 | |
8UZO
 
 | |
8UZM
 
 | |
8USI
 
 | |
8UZI
 
 | |
8USL
 
 | |
8UQO
 
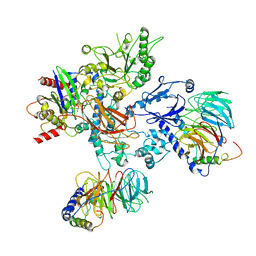 | | PLCb3-Gbg-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8USH
 
 | |
8USF
 
 | |
8USJ
 
 | |
7MXR
 
 | |
7MXS
 
 | |
8USG
 
 | |
7MXU
 
 | |
7MXV
 
 | |
7MXT
 
 | |
7MXQ
 
 | |
8UQN
 
 | | PLCb3-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7MXW
 
 | |
8UZK
 
 | |
7MXX
 
 | |
6SGR
 
 | | Cryo-EM structure of Escherichia coli AcrBZ and DARPin in Saposin A-nanodisc with cardiolipin | | Descriptor: | DARPin, Multidrug efflux pump accessory protein AcrZ, Multidrug efflux pump subunit AcrB | | Authors: | Szewczak-Harris, A, Du, D, Newman, C, Neuberger, A, Luisi, B.F. | | Deposit date: | 2019-08-05 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Interactions of a Bacterial RND Transporter with a Transmembrane Small Protein in a Lipid Environment.
Structure, 28, 2020
|
|
8UR6
 
 | |
