6X50
 
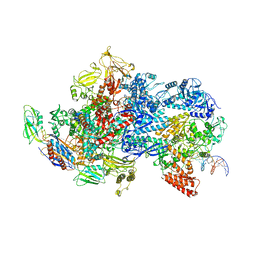 | | Mfd-bound E.coli RNA polymerase elongation complex - V state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewelyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
4RQF
 
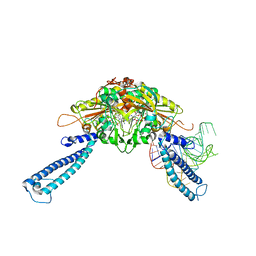 | | human Seryl-tRNA synthetase dimer complexed with one molecule of tRNAsec | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SERINE, Serine--tRNA ligase, ... | | Authors: | Xie, W, Wang, C, Guo, Y, Tian, Q, Jia, Q. | | Deposit date: | 2014-11-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.503 Å) | | Cite: | SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis.
Nucleic Acids Res., 43, 2015
|
|
4RQE
 
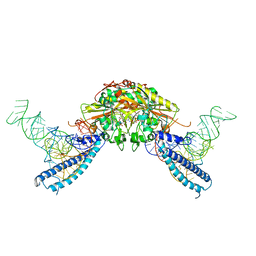 | | human Seryl-tRNA synthetase dimer complexed with two molecules of tRNAsec | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SERINE, Serine--tRNA ligase, ... | | Authors: | Xie, W, Wang, C, Guo, Y, Tian, Q, Jia, Q. | | Deposit date: | 2014-11-03 | | Release date: | 2015-11-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis.
Nucleic Acids Res., 43, 2015
|
|
6X2F
 
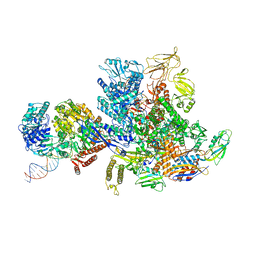 | | Mfd-bound E.coli RNA polymerase elongation complex - L2 state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X2N
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - I state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X26
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - L1 state | | Descriptor: | DNA (64-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X4W
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - III state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X4Y
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - IV state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X43
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-22 | | Release date: | 2021-02-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
3TYV
 
 | |
7AHO
 
 | | RUVBL1-RUVBL2 heterohexameric ring after binding of RNA helicase DHX34 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Lopez-Perrote, A, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2020-09-25 | | Release date: | 2020-11-25 | | Method: | ELECTRON MICROSCOPY (4.18 Å) | | Cite: | Regulation of RUVBL1-RUVBL2 AAA-ATPases by the nonsense-mediated mRNA decay factor DHX34, as evidenced by Cryo-EM.
Elife, 9, 2020
|
|
7NVX
 
 | | TFIIH in a post-translocated state (with ADP-BeF3) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, CDK-activating kinase assembly factor MAT1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
7NVW
 
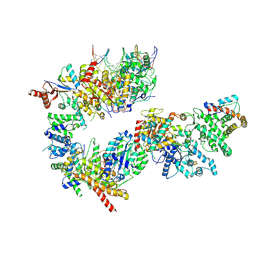 | | TFIIH in a pre-translocated state (without ADP-BeF3) | | Descriptor: | CDK-activating kinase assembly factor MAT1, General transcription and DNA repair factor IIH helicase subunit XPB, General transcription factor IIE subunit 1, ... | | Authors: | Aibara, S, Schilbach, S, Cramer, P. | | Deposit date: | 2021-03-16 | | Release date: | 2021-05-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structures of mammalian RNA polymerase II pre-initiation complexes.
Nature, 594, 2021
|
|
6H9R
 
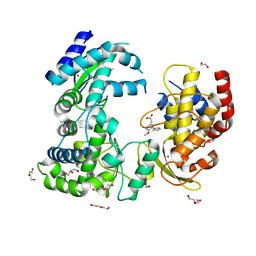 | | Dengue-RdRp3-inhibitor complex soaking | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
6NPW
 
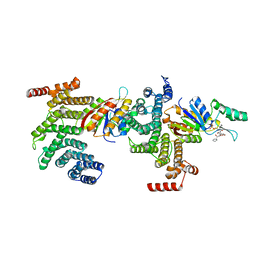 | | SSu72/Sympk in complex with Ser2/Ser5 phosphorylated peptide | | Descriptor: | PHOSPHATE ION, Ser2/Ser5 phosphorylated peptide, Ssu72 ortholog, ... | | Authors: | Irani, S, Zhang, Y. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.486 Å) | | Cite: | Structural determinants for accurate dephosphorylation of RNA polymerase II by its cognate C-terminal domain (CTD) phosphatase during eukaryotic transcription.
J.Biol.Chem., 294, 2019
|
|
6H80
 
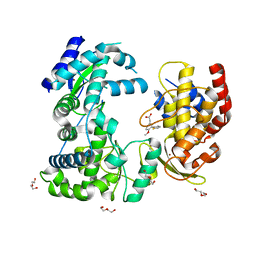 | | Dengue-RdRp3-inhibitor complex co-crystallisation | | Descriptor: | 2-(4-methoxy-3-thiophen-2-yl-phenyl)ethanoic acid, DI(HYDROXYETHYL)ETHER, Genome polyprotein, ... | | Authors: | Talapatra, S.K, Kozielski, F. | | Deposit date: | 2018-07-31 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Development and validation of RdRp Screen, a crystallization screen for viral RNA-dependent RNA polymerases.
Biol Open, 8, 2019
|
|
8RE4
 
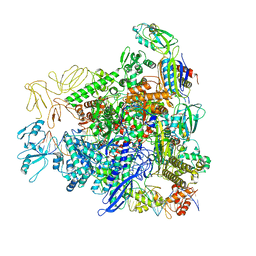 | |
8RED
 
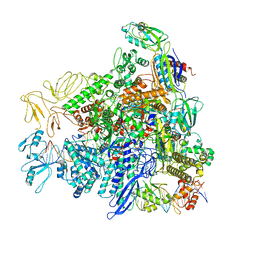 | |
8REC
 
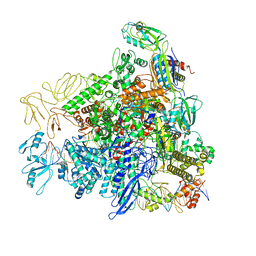 | |
8REE
 
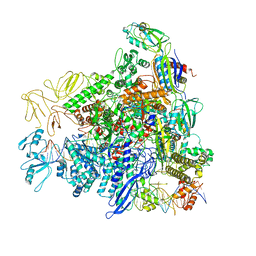 | |
6FLP
 
 | |
8KDC
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in monomeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
8KDB
 
 | | Cryo-EM structure of the human parainfluenza virus hPIV3 L-P polymerase in dimeric form | | Descriptor: | MAGNESIUM ION, Phosphoprotein, RNA-directed RNA polymerase L, ... | | Authors: | Xie, J, Wang, L, Zhai, G, Wu, D, Lin, Z, Wang, M, Yan, X, Gao, L, Huang, X, Fearns, R, Chen, S. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural basis for dimerization of a paramyxovirus polymerase complex.
Nat Commun, 15, 2024
|
|
8REB
 
 | |
8REA
 
 | |
