7Q5Y
 
 | |
4I3J
 
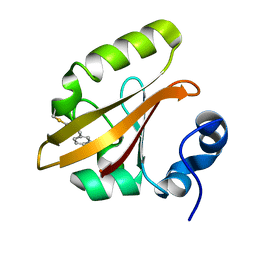 | | Structures of PR1 intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected AT 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
2MP4
 
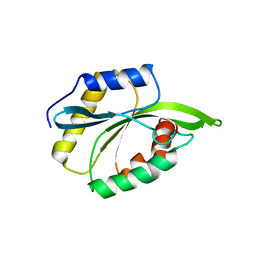 | | Solution Structure of ADF like UNC-60A Protein of Caenorhabditis elegans | | Descriptor: | Actin-depolymerizing factor 1, isoforms a/b | | Authors: | Shukla, V, Yadav, R, Kabra, A, Kumar, D, Ono, S, Arora, A. | | Deposit date: | 2014-05-11 | | Release date: | 2014-06-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR Structure and Backbone dynamics of ADF like UNC-60A protein from Caenorhabditis elegans: its divergence from conventional ADF/cofilin
To be Published
|
|
7OK2
 
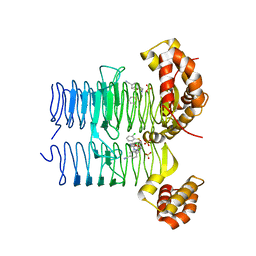 | | Crystal structure of Pseudomonas aeruginosa LpxA in complex with compound 3 | | Descriptor: | Acyl-[acyl-carrier-protein]-UDP-N-acetylglucosamine O-acyltransferase, SULFATE ION, ~{N}-[(5-azanyl-1,3,4-oxadiazol-2-yl)methyl]-2-(2-chlorophenyl)sulfanyl-~{N}-[(4-cyanophenyl)methyl]ethanamide | | Authors: | Ryan, M.D, Parkes, A.L, Southey, M, Andersen, O.A, Zahn, M, Barker, J, DeJonge, B.L.M. | | Deposit date: | 2021-05-17 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Discovery of Novel UDP- N -Acetylglucosamine Acyltransferase (LpxA) Inhibitors with Activity against Pseudomonas aeruginosa .
J.Med.Chem., 64, 2021
|
|
8FYH
 
 | | G4 RNA-mediated PRC2 dimer | | Descriptor: | G4 RNA, Histone-binding protein RBBP4, Histone-lysine N-methyltransferase EZH2, ... | | Authors: | Song, J, Kasinath, V. | | Deposit date: | 2023-01-26 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for inactivation of PRC2 by G-quadruplex RNA.
Science, 381, 2023
|
|
1O5O
 
 | |
2ZZ6
 
 | |
4QVP
 
 | | yCP beta5-M45T mutant in complex with bortezomib | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, N-[(1R)-1-(DIHYDROXYBORYL)-3-METHYLBUTYL]-N-(PYRAZIN-2-YLCARBONYL)-L-PHENYLALANINAMIDE, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
5FVB
 
 | | CRYSTAL STRUCTURE OF PHORMIDIUM C-PHYCOERYTHRIN AT PH 5.0 | | Descriptor: | C-PHYCOERYTHRIN ALPHA SUBUNIT, C-PHYCOERYTHRIN BETA SUBUNIT, GLYCEROL, ... | | Authors: | Kumar, V, Sonani, R.R, Sharma, M, Gupta, G.D, Madamwar, D. | | Deposit date: | 2016-02-05 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal Structure Analysis of C-Phycoerythrin from Marine Cyanobacterium Phormidium Sp. A09Dm.
Photosynth.Res., 129, 2016
|
|
4HGF
 
 | | Crystal structure of P450 BM3 5F5K heme domain variant complexed with styrene | | Descriptor: | Bifunctional P-450/NADPH-P450 reductase, CHLORIDE ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Shehzad, A, Panneerselvam, S, Bocola, M, Mueller-Dieckmann, J, Wilmanns, M, Schwaneberg, U. | | Deposit date: | 2012-10-08 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | P450 BM3 crystal structures reveal the role of the charged surface residue Lys/Arg184 in inversion of enantioselective styrene epoxidation.
Chem.Commun.(Camb.), 49, 2013
|
|
1NOW
 
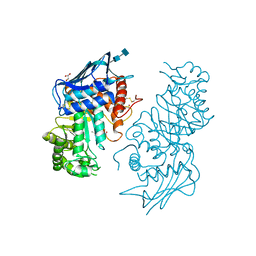 | | Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
4QWK
 
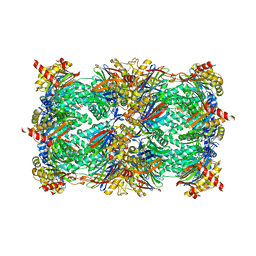 | | yCP beta5-A49T-A50V-double mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4HCY
 
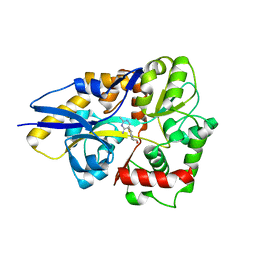 | | Structure of a eukaryotic thiaminase-I bound to the thiamin analogue 3-deazathiamin | | Descriptor: | 2-{4-[(4-amino-2-methylpyrimidin-5-yl)methyl]-3-methylthiophen-2-yl}ethanol, thiaminase-I | | Authors: | Kreinbring, C.A, Hubbard, P.A, Leeper, F.J, Hawksley, D, Petsko, G.A, Ringe, D. | | Deposit date: | 2012-10-01 | | Release date: | 2013-10-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure of a eukaryotic thiaminase I.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
3AJA
 
 | |
4QZ1
 
 | | yCP beta5-M45T mutant in complex with the epoxyketone inhibitor ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-27 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
6XGR
 
 | | YSD1 major tail protein | | Descriptor: | YSD1_22 major tail protein | | Authors: | Hardy, J.M, Dunstan, R, Venugopal, H, Lithgow, T.J, Coulibaly, F.J. | | Deposit date: | 2020-06-17 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | The architecture and stabilisation of flagellotropic tailed bacteriophages.
Nat Commun, 11, 2020
|
|
4R18
 
 | | Ligand-induced Lys33-Thr1 crosslinking at subunit beta5 of the yeast 20S proteasome | | Descriptor: | ALPHA-AMINOBUTYRIC ACID, MAGNESIUM ION, PROTEASOME SUBUNIT ALPHA TYPE-1, ... | | Authors: | Dubiella, C, Cui, H, Gersch, M, Brouwer, A.J, Sieber, S.A, Krueger, A, Liskamp, R, Groll, M. | | Deposit date: | 2014-08-04 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Selective inhibition of the immunoproteasome by ligand-induced crosslinking of the active site.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1QVF
 
 | | Structure of a deacylated tRNA minihelix bound to the E site of the large ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S ribosomal rna, 50S RIBOSOMAL PROTEIN L10E, 50S ribosomal protein L13P, ... | | Authors: | Schmeing, T.M, Moore, P.B, Steitz, T.A. | | Deposit date: | 2003-08-27 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of deacylated tRNA mimics bound to the E site of the large ribosomal subunit
RNA, 9, 2003
|
|
4QV4
 
 | | yCP beta5-M45T mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
7OKN
 
 | |
4QW6
 
 | | yCP beta5-M45V mutant in complex with carfilzomib | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Huber, E.M, Heinemeyer, W, Groll, M. | | Deposit date: | 2014-07-16 | | Release date: | 2015-02-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bortezomib-Resistant Mutant Proteasomes: Structural and Biochemical Evaluation with Carfilzomib and ONX 0914.
Structure, 23, 2015
|
|
4N04
 
 | |
6X6K
 
 | | Cryo-EM Structure of the Helicobacter pylori dCag3 OMC | | Descriptor: | Cag pathogenicity island protein, Cag pathogenicity island protein (Cag7), Type IV secretion system apparatus protein CagX | | Authors: | Sheedlo, M.J, Chung, J.M, Sawhney, N, Durie, C.L, Cover, T.L, Ohi, M.D, Lacy, D.B. | | Deposit date: | 2020-05-28 | | Release date: | 2020-10-07 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM reveals species-specific components within the Helicobacter pylori Cag type IV secretion system core complex.
Elife, 9, 2020
|
|
4QY1
 
 | | Structure of H10 from human-infecting H10N8 in complex with avian receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, M, Zhang, W, Qi, J, Wang, F, Zhou, J, Bi, Y, Wu, Y, Sun, H, Liu, J, Huang, C, Li, X, Yan, J, Shu, Y, Shi, Y, Gao, G.F. | | Deposit date: | 2014-07-23 | | Release date: | 2015-01-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | Structural basis for preferential avian receptor binding by the human-infecting H10N8 avian influenza virus
Nat Commun, 6, 2015
|
|
1Q6Q
 
 | | Structure of 3-keto-L-gulonate 6-phosphate decarboxylase with bound xylitol 5-phosphate | | Descriptor: | 3-keto-L-gulonate 6-phosphate decarboxylase, L-XYLITOL 5-PHOSPHATE, MAGNESIUM ION | | Authors: | Wise, E.L, Yew, W.S, Gerlt, J.A, Rayment, I. | | Deposit date: | 2003-08-13 | | Release date: | 2003-10-28 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.695 Å) | | Cite: | Structural Evidence for a 1,2-Enediolate Intermediate in the Reaction Catalyzed by 3-Keto-l-Gulonate 6-Phosphate Decarboxylase, a Member of the Orotidine 5'-Monophosphate Decarboxylase Suprafamily
Biochemistry, 42, 2003
|
|
