3Q2W
 
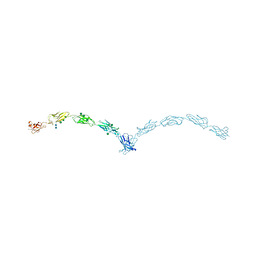 | | Crystal structure of mouse N-cadherin ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
4B1C
 
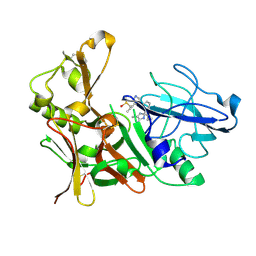 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-cyclopropyl-5-methyl-2-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-2H-imidazol-4-amine, BETA-SECRETASE 1, DIMETHYL SULFOXIDE | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
1GVZ
 
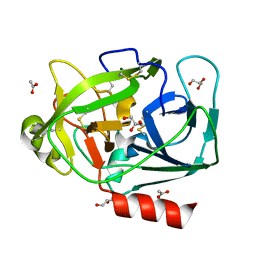 | | Prostate Specific Antigen (PSA) from stallion seminal plasma | | Descriptor: | ACETATE ION, GLYCEROL, KALLIKREIN-1E2 | | Authors: | Carvalho, A.L, Sanz, L, Barettino, D, Romero, A, Calvete, J.J, Romao, M.J. | | Deposit date: | 2002-02-28 | | Release date: | 2002-09-12 | | Last modified: | 2011-09-21 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Crystal Structure of a Prostate Kallikrein Isolated from Stallion Seminal Plasma: A Homologue of Human Psa
J.Mol.Biol., 322, 2002
|
|
7XMP
 
 | |
4DY0
 
 | | Crystal structure of native protease nexin-1 with heparin | | Descriptor: | 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid, GLYCEROL, Glia-derived nexin, ... | | Authors: | Huntington, J.A, Li, W. | | Deposit date: | 2012-02-28 | | Release date: | 2012-08-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of protease nexin-1 in complex with heparin and thrombin suggest a 2-step recognition mechanism.
Blood, 120, 2012
|
|
2Z3U
 
 | | Crystal Structure of Chromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 1,2-ETHANEDIOL, 3,4-DI-1H-INDOL-3-YL-1H-PYRROLE-2,5-DICARBOXYLIC ACID, Cytochrome P450, ... | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3PI3
 
 | | Crystallographic Structure of HbII-oxy from Lucina pectinata at pH 5.0 | | Descriptor: | Hemoglobin II, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gavira, J.A, Nieves-Marrero, C.A, Ruiz-Martinez, C.R, Estremera-Andujar, R.A, Lopez-Garriga, J, Garcia-Ruiz, J.M. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | pH-dependence crystallographic studies of the oxygen carrier hemoglobin II from Lucina pectinata
To be Published
|
|
3PI0
 
 | | Endothiapepsin in complex with a fragment | | Descriptor: | DIMETHYL SULFOXIDE, Endothiapepsin, GLYCEROL, ... | | Authors: | Koester, H, Heine, A, Klebe, G. | | Deposit date: | 2010-11-05 | | Release date: | 2011-11-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A small nonrule of 3 compatible fragment library provides high hit rate of endothiapepsin crystal structures with various fragment chemotypes.
J.Med.Chem., 54, 2011
|
|
3PI9
 
 | | Site-specific Glycosylation of Hemoglobin Utilizing Oxime Ligation Chemistry as a Viable Alternative to PEGylation | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Bhatt, V.S, Styslinger, T.J, Zhang, N, Wang, P.G, Palmer, A.F. | | Deposit date: | 2010-11-05 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: |
|
|
2Z1I
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
3PMJ
 
 | | Bovine trypsin variant X(tripleIle227) in complex with small molecule inhibitor | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cationic trypsin, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-11-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4B7R
 
 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Liu, J, van der Vries, E, Vachieri, S.G, Xiong, X, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
3PPV
 
 | | Crystal structure of an engineered VWF A2 domain (N1493C and C1670S) | | Descriptor: | CALCIUM ION, SULFATE ION, von Willebrand factor | | Authors: | Zhou, M, Dong, X, Zhong, C, Ding, J. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A novel calcium-binding site of von Willebrand factor A2 domain regulates its cleavage by ADAMTS13
Blood, 117, 2011
|
|
2Z3T
 
 | | Crystal Structure of Substrate Free Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome P450, IMIDAZOLE, ... | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2007-06-06 | | Release date: | 2007-07-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
4B78
 
 | | Aminoimidazoles as BACE-1 Inhibitors: From De Novo Design to Ab- lowering in Brain | | Descriptor: | (3R,5R)-3-methoxy-5-(4-methoxyphenyl)-5-(3-pyridin-3-ylphenyl)-3,4-dihydropyrrol-2-amine, BETA-SECRETASE 1 | | Authors: | Gravenfors, Y, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, Berg, S, Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Rahm, F. | | Deposit date: | 2012-08-16 | | Release date: | 2013-06-26 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Core Refinement Toward Permeable Beta-Secretase (Bace-1) Inhibitors with Low Herg Activity.
J.Med.Chem., 56, 2013
|
|
3PSJ
 
 | |
2Z57
 
 | | Crystal structure of G56E-propeptide:S324A-subtilisin complex | | Descriptor: | CALCIUM ION, Tk-subtilisin, ZINC ION | | Authors: | Pulido, M.A, Tanaka, S, Sringiew, C, You, D.J, Matsumura, H, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-06-29 | | Release date: | 2008-01-01 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Requirement of left-handed glycine residue for high stability of the Tk-subtilisin propeptide as revealed by mutational and crystallographic analyses
J.Mol.Biol., 374, 2007
|
|
4B00
 
 | | Design and Synthesis of BACE1 Inhibitors with In Vivo Brain Reduction of beta-Amyloid Peptides (COMPOUND (R)-41) | | Descriptor: | 5-{(1R)-3-amino-4-fluoro-1-[3-(5-prop-1-yn-1-ylpyridin-3-yl)phenyl]-1H-isoindol-1-yl}-1-ethyl-3-methylpyridin-2(1H)-one, ACETATE ION, BETA-SECRETASE 1 | | Authors: | Swahn, B.M, Kolmodin, K, Karlstrom, S, von Berg, S, Soderman, P, Holenz, J, Berg, S, Lindstrom, J, Sundstrom, M, Turek, D, Kihlstrom, J, Slivo, C, Andersson, L, Pyring, D, Ohberg, L, Kers, A, Bogar, K, Bergh, M, Olsson, L.L, Janson, J, Eketjall, S, Georgievska, B, Jeppsson, F, Falting, J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-10-17 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Design and synthesis of beta-site amyloid precursor protein cleaving enzyme (BACE1) inhibitors with in vivo brain reduction of beta-amyloid peptides.
J. Med. Chem., 55, 2012
|
|
3PRK
 
 | | INHIBITION OF PROTEINASE K BY METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE. AN X-RAY STUDY AT 2.2-ANGSTROMS RESOLUTION | | Descriptor: | CALCIUM ION, METHOXYSUCCINYL-ALA-ALA-PRO-ALA-CHLOROMETHYL KETONE, PROTEINASE K | | Authors: | Wolf, W.M, Bajorath, J, Mueller, A, Raghunathan, S, Singh, T.P, Hinrichs, W, Saenger, W. | | Deposit date: | 1991-08-07 | | Release date: | 1994-01-31 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibition of proteinase K by methoxysuccinyl-Ala-Ala-Pro-Ala-chloromethyl ketone. An x-ray study at 2.2-A resolution.
J.Biol.Chem., 266, 1991
|
|
4DC4
 
 | | Lysozyme Trimer | | Descriptor: | CHLORIDE ION, GLYCEROL, Lysozyme C, ... | | Authors: | Sharma, P, Ashish | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Characterization of heat induced spherulites of lysozyme reveals new insight on amyloid initiation
Sci Rep, 6, 2016
|
|
4JP1
 
 | | Mg2+ bound structure of Vibrio Cholerae CheY3 | | Descriptor: | Chemotaxis protein CheY, MAGNESIUM ION | | Authors: | Biswas, M, Dasgupta, J, Sen, U. | | Deposit date: | 2013-03-19 | | Release date: | 2014-04-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Conformational barrier of CheY3 and inability of CheY4 to bind FliM control the flagellar motor action in Vibrio cholerae
Plos One, 8, 2013
|
|
4J62
 
 | |
2Z1J
 
 | | Crystal structure of E.coli RNase HI surface charged mutant(Q4R/T40E/Q72H/Q76K/Q80E/T92K/Q105K/Q113R/Q115K/N143K/T145K) | | Descriptor: | Ribonuclease HI | | Authors: | You, D.J, Fukuchi, S, Nishikawa, K, Koga, Y, Takano, K, Kanaya, S. | | Deposit date: | 2007-05-08 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Protein Thermostabilization Requires a Fine-tuned Placement of Surface-charged Residues
J.Biochem.(Tokyo), 142, 2007
|
|
4JP8
 
 | | Crystal structure of Pro-F17H/S324A | | Descriptor: | CALCIUM ION, Tk-subtilisin | | Authors: | Yuzaki, K, You, D.J, Uehara, R, Koga, Y, Kanaya, S. | | Deposit date: | 2013-03-19 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Increase in activation rate of Pro-Tk-subtilisin by a single nonpolar-to-polar amino acid substitution at the hydrophobic core of the propeptide domain
Protein Sci., 22, 2013
|
|
4J69
 
 | | Crystal structure of Ribonuclease A soaked in 50% S,R,S-bisfuranol: One of twelve in MSCS set | | Descriptor: | (3S,3aR,6aS)-hexahydrofuro[2,3-b]furan-3-ol, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kearney, B.M, Dechene, M, Swartz, P.D, Mattos, C. | | Deposit date: | 2013-02-11 | | Release date: | 2014-01-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.892 Å) | | Cite: | DRoP: A program for analysis of water structure on protein surfaces
to be published
|
|
