3ZUW
 
 | | Photosynthetic Reaction Centre Mutant with TYR L128 replaced with HIS | | Descriptor: | BACTERIOCHLOROPHYLL A, BACTERIOPHEOPHYTIN A, FE (III) ION, ... | | Authors: | Gibasiewicz, K, Pajzderska, M, Potter, J.A, Fyfe, P.K, Dobek, A, Brettel, K, Jones, M.R. | | Deposit date: | 2011-07-20 | | Release date: | 2011-11-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Mechanism of Recombination of the P(+)H(A)(-) Radical Pair in Mutant Rhodobacter Sphaeroides Reaction Centers with Modified Free Energy Gaps between P(+)B(A)(-) and P(+)H(A)(-).
J Phys Chem B, 115, 2011
|
|
4B7G
 
 | | Structure of a bacterial catalase | | Descriptor: | CATALASE, CHLORIDE ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
4B7H
 
 | | Structure of a highdose liganded bacterial catalase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CATALASE, CHLORIDE ION, ... | | Authors: | Gumiero, A, Walsh, M. | | Deposit date: | 2012-08-20 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | A Structural and Dynamic Investigation of the Inhibition of Catalase by Nitric Oxide.
Org.Biomol.Chem., 11, 2013
|
|
1C0B
 
 | |
6DX0
 
 | | Hermes transposase deletion dimer complex with (A/T) DNA | | Descriptor: | DNA (25-MER), DNA (5'-D(*AP*GP*AP*GP*AP*AP*CP*AP*AP*CP*AP*AP*CP*AP*AP*G)-3'), DNA (5'-D(*GP*CP*GP*TP*GP*AP*A)-3'), ... | | Authors: | Dyda, F, Voth, A.R, Hickman, A.B. | | Deposit date: | 2018-06-28 | | Release date: | 2018-09-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural insights into the mechanism of double strand break formation by Hermes, a hAT family eukaryotic DNA transposase.
Nucleic Acids Res., 46, 2018
|
|
6E5Z
 
 | |
6KE0
 
 | | Crystal structure of PDE10A in complex with a triazolopyrimidine inhibitor | | Descriptor: | 2-(5,7-dimethyl-[1,2,4]triazolo[1,5-a]pyrimidin-2-yl)-1-[(2S)-2-methyl-1,2-dihydroimidazo[1,2-a]benzimidazol-3-yl]ethanone, MAGNESIUM ION, ZINC ION, ... | | Authors: | Amano, Y, Honbou, K. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Synthesis, SAR study, and biological evaluation of novel 2,3-dihydro-1H-imidazo[1,2-a]benzimidazole derivatives as phosphodiesterase 10A inhibitors.
Bioorg.Med.Chem., 27, 2019
|
|
1A61
 
 | | THROMBIN COMPLEXED WITH A BETA-MIMETIC THIAZOLE-CONTAINING INHIBITOR | | Descriptor: | (1S,7S)-7-amino-7-benzyl-N-{(1S)-4-carbamimidamido-1-[(S)-hydroxy(1,3-thiazol-2-yl)methyl]butyl}-8-oxohexahydro-1H-pyra zolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Matthews, J.H, Zhang, E, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-03-05 | | Release date: | 1998-06-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
4NXN
 
 | | Crystal Structure of the 30S ribosomal subunit from a GidB (RsmG) mutant of Thermus thermophilus (HB8), bound with streptomycin | | Descriptor: | 16S rRNA, MAGNESIUM ION, STREPTOMYCIN, ... | | Authors: | Demirci, H, Murphy IV, F, Murphy, E, Gregory, S.T, Dahlberg, A.E, Jogl, G. | | Deposit date: | 2013-12-09 | | Release date: | 2014-05-21 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (3.544 Å) | | Cite: | A structural basis for streptomycin resistance
To be Published
|
|
5EAQ
 
 | |
4BYJ
 
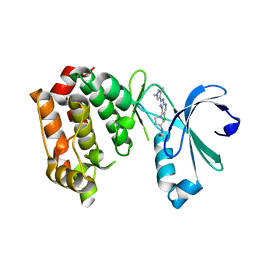 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-(1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4ZE9
 
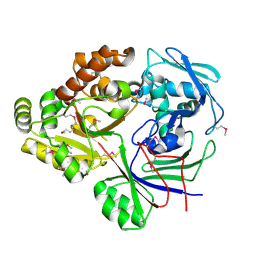 | | Se-PBP AccA from A. tumefaciens C58 in complex with agrocinopine A | | Descriptor: | 2-O-phosphono-alpha-L-arabinopyranose, ABC transporter substrate-binding protein, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | El Sahili, A, Guimaraes, B.G, Morera, S. | | Deposit date: | 2015-04-20 | | Release date: | 2015-08-19 | | Last modified: | 2023-06-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Pyranose-2-Phosphate Motif Is Responsible for Both Antibiotic Import and Quorum-Sensing Regulation in Agrobacterium tumefaciens.
Plos Pathog., 11, 2015
|
|
5OVG
 
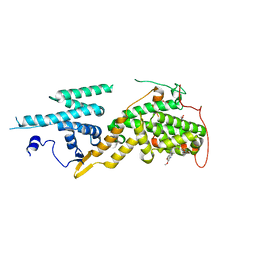 | | Ras guanine nucleotide exchange factor SOS1 (Rem-cdc25) in complex with small molecule inhibitor compound 18 | | Descriptor: | 1,2-ETHANEDIOL, Son of sevenless homolog 1, ~{N}-[(1~{R})-1-[5-(6,7-dihydro-5~{H}-pyrrolo[1,2-a]imidazol-3-yl)thiophen-2-yl]ethyl]-6,7-dimethoxy-2-methyl-quinazolin-4-amine | | Authors: | Hillig, R.C, Sautier, B, Schroeder, J, Moosmayer, D, Hilpmann, A, Stegmann, C.M, Briem, H, Boemer, U, Weiske, J, Badock, V, Petersen, K, Kahmann, J, Wegener, D, Bohnke, N, Eis, K, Graham, K, Wortmann, L, von Nussbaum, F, Bader, B. | | Deposit date: | 2017-08-28 | | Release date: | 2019-02-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of potent SOS1 inhibitors that block RAS activation via disruption of the RAS-SOS1 interaction.
Proc. Natl. Acad. Sci. U.S.A., 116, 2019
|
|
4AQR
 
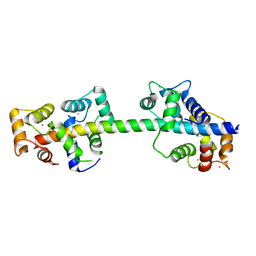 | | Crystal structure of calmodulin in complex with the regulatory domain of a plasma-membrane Ca2+-ATPase | | Descriptor: | CALCIUM ION, CALCIUM-TRANSPORTING ATPASE 8, PLASMA MEMBRANE-TYPE, ... | | Authors: | Tidow, H, Poulsen, L.R, Andreeva, A, Hein, K.L, Palmgren, M.G, Nissen, P. | | Deposit date: | 2012-04-19 | | Release date: | 2012-10-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Bimodular Mechanism of Calcium Control in Eukaryotes
Nature, 491, 2012
|
|
1BCJ
 
 | |
6ERJ
 
 | | Self-complemented FimA subunit from Salmonella enterica | | Descriptor: | ACETIC ACID, Type-1 fimbrial protein, a chain | | Authors: | Zyla, D.S, Prota, A, Capitani, G, Glockshuber, R. | | Deposit date: | 2017-10-18 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Alternative folding to a monomer or homopolymer is a common feature of the type 1 pilus subunit FimA from enteroinvasive bacteria.
J.Biol.Chem., 294, 2019
|
|
4ACF
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS GLUTAMINE SYNTHETASE IN COMPLEX WITH IMIDAZOPYRIDINE INHIBITOR ((4-(6-BROMO-3-(BUTYLAMINO)IMIDAZO(1,2-A)PYRIDIN-2-YL)PHENOXY) ACETIC ACID) AND L-METHIONINE-S-SULFOXIMINE PHOSPHATE. | | Descriptor: | CHLORIDE ION, GLUTAMINE SYNTHETASE 1, L-METHIONINE-S-SULFOXIMINE PHOSPHATE, ... | | Authors: | Nilsson, M.T, Mowbray, S.L. | | Deposit date: | 2011-12-15 | | Release date: | 2012-10-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Synthesis, Biological Evaluation and X-Ray Crystallographic Studies of Imidazo(1,2-A)Pyridine-Based Mycobacterium Tuberculosis Glutamine Synthetase Inhibitors
Medchemcomm, 3, 2012
|
|
1B2V
 
 | | HEME-BINDING PROTEIN A | | Descriptor: | CALCIUM ION, PROTEIN (HEME-BINDING PROTEIN A), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Arnoux, P, Haser, R, Izadi, N, Lecroisey, A, Wandersma, N.C, Czjzek, M. | | Deposit date: | 1998-12-01 | | Release date: | 1999-06-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of HasA, a hemophore secreted by Serratia marcescens.
Nat.Struct.Biol., 6, 1999
|
|
7YML
 
 | | Structure of photosynthetic LH1-RC super-complex of Rhodobacter capsulatus | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, BACTERIOCHLOROPHYLL A, ... | | Authors: | Tani, K, Kanno, R, Ji, X.-C, Satoh, I, Kobayashi, Y, Nagashima, K.V.P, Hall, M, Yu, L.-J, Kimura, Y, Mizoguchi, A, Humbel, B.M, Madigan, M.T, Wang-Otomo, Z.-Y. | | Deposit date: | 2022-07-28 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Rhodobacter capsulatus forms a compact crescent-shaped LH1-RC photocomplex.
Nat Commun, 14, 2023
|
|
7VVR
 
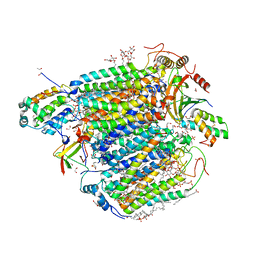 | | Bovine cytochrome c oxidese in CN-bound mixed valence state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-08 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
7ZG8
 
 | |
1A46
 
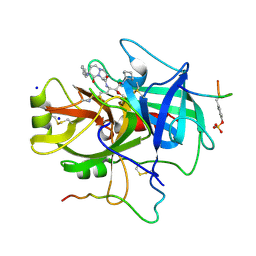 | | THROMBIN COMPLEXED WITH HIRUGEN AND A BETA-STRAND MIMETIC INHIBITOR | | Descriptor: | (1S,7S)-7-amino-N-[(2R,3S)-7-amino-1-(cyclohexylamino)-2-hydroxy-1-oxoheptan-3-yl]-7-benzyl-8-oxohexahydro-1H-pyrazolo[1,2-a]pyridazine-1-carboxamide, ALPHA-THROMBIN (LARGE SUBUNIT), ALPHA-THROMBIN (SMALL SUBUNIT), ... | | Authors: | St Charles, R, Matthews, J.H, Zhang, E, Tulinsky, A, Kahn, M. | | Deposit date: | 1998-02-11 | | Release date: | 1998-05-27 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Bound structures of novel P3-P1' beta-strand mimetic inhibitors of thrombin.
J.Med.Chem., 42, 1999
|
|
7VUW
 
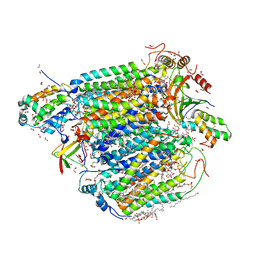 | | Bovine heart cytochrome c oxidase in the cyanide-bound fully oxidized state at 50 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Shimada, A, Tsukihara, T. | | Deposit date: | 2021-11-04 | | Release date: | 2022-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic cyanide-probing for cytochrome c oxidase reveals structural bases suggesting that a putative proton transfer H-pathway pumps protons.
J.Biol.Chem., 299, 2023
|
|
4BYI
 
 | | Aurora A kinase bound to a highly selective imidazopyridine inhibitor | | Descriptor: | (S)-N-((1-(6-chloro-2-(1,3-dimethyl-1H-pyrazol-4-yl)-3H-imidazo[4,5-b]pyridin-7-yl)pyrrolidin-3-yl)methyl)acetamide, AURORA KINASE A | | Authors: | Joshi, A, Kosmopoulou, M, Bayliss, R. | | Deposit date: | 2013-07-19 | | Release date: | 2013-11-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Aurora Isoform Selectivity: Design and Synthesis of Imidazo[4,5-B]Pyridine Derivatives as Highly Selective Inhibitors of Aurora-A Kinase in Cells.
J.Med.Chem., 56, 2013
|
|
4C9C
 
 | | Crystal Structure of the Strawberry Pathogenesis-Related 10 (PR-10) Fra a 1E protein (Form A) | | Descriptor: | GLYCEROL, MAJOR STRAWBERRY ALLERGEN FRA A 1-E, SULFATE ION | | Authors: | Casanal, A, Zander, U, Valpuesta, V, Marquez, J.A. | | Deposit date: | 2013-10-02 | | Release date: | 2013-10-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Strawberry Pathogenesis-Related 10 (Pr-10) Fra a Proteins Control Flavonoid Biosynthesis by Binding to Metabolic Intermediates.
J.Biol.Chem., 288, 2013
|
|
