6GQQ
 
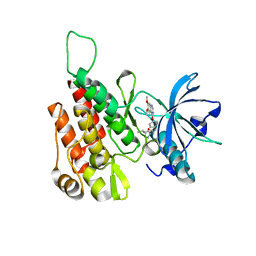 | | Crystal structure of human KDR (VEGFR2) kinase domain in complex with AZD3229-analogue (compound 35) | | Descriptor: | Vascular endothelial growth factor receptor 2, ~{N}-[4-(6,7-dimethoxyquinazolin-4-yl)oxyphenyl]-2-(4-propan-2-yl-1,2,3-triazol-1-yl)ethanamide | | Authors: | Schimpl, M, Hardy, C.J, Ogg, D.J, Overman, R.C, Packer, M.J, Kettle, J.G, Anjum, R, Barry, E, Bhavsar, D, Brown, C, Campbell, A, Goldberg, K, Grondine, M, Guichard, S, Hunt, T, Jones, O, Li, X, Moleva, O, Pearson, S, Shao, W, Smith, A, Smith, J, Stead, D, Stokes, S, Tucker, M, Ye, Y. | | Deposit date: | 2018-06-07 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Discovery of N-(4-{[5-Fluoro-7-(2-methoxyethoxy)quinazolin-4-yl]amino}phenyl)-2-[4-(propan-2-yl)-1 H-1,2,3-triazol-1-yl]acetamide (AZD3229), a Potent Pan-KIT Mutant Inhibitor for the Treatment of Gastrointestinal Stromal Tumors.
J. Med. Chem., 61, 2018
|
|
8AHW
 
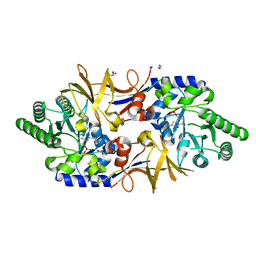 | | Structure of DCS-resistant variant D322N of alanine racemase from Mycobacterium tuberculosis | | Descriptor: | 1,2-ETHANEDIOL, Alanine racemase, GLYCEROL | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-07-22 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
6WF5
 
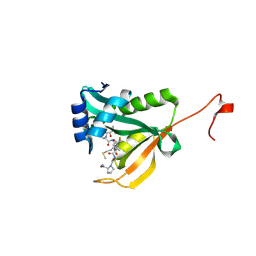 | | Crystal structure of human Naa50 in complex with a truncated cofactor derived inhibitor (compound 2) | | Descriptor: | (2R)-2-hydroxy-3,3-dimethyl-N-{3-oxo-3-[(2-sulfanylethyl)amino]propyl}butanamide, ACE-MET-LEU-GLY-PRO-NH2, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFG
 
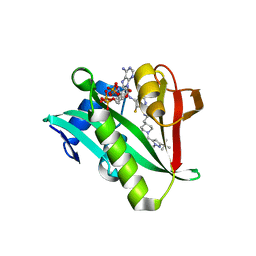 | | Crystal structure of human Naa50 in complex with an inhibitor (compound 3) identified using DNA encoded library technology | | Descriptor: | (2S)-N-[(2S)-3-[1-(3-tert-butyl-1-methyl-1H-pyrazole-5-carbonyl)piperidin-4-yl]-1-(methylamino)-1-oxopropan-2-yl]-6-oxopiperidine-2-carboxamide, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFO
 
 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4b) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5R)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
1BQ2
 
 | | E. COLI THYMIDYLATE SYNTHASE MUTANT N177A | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Reyes, C.L, Sage, C.R, Rutenber, E.E, Finer-Moore, J.S, Stroud, R.M. | | Deposit date: | 1998-08-20 | | Release date: | 1999-04-27 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inactivity of N229A thymidylate synthase due to water-mediated effects: isolating a late stage in methyl transfer.
J.Mol.Biol., 284, 1998
|
|
8B8H
 
 | | Structure of DCS-resistant variant D322N of alanine racemase from M. tuberculosis in complex with DCS | | Descriptor: | (~{E})-[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylidene-[(4~{R})-3-oxidanylidene-1,2-oxazolidin-4-yl]azanium, 1,2-ETHANEDIOL, Alanine racemase, ... | | Authors: | de Chiara, C, Prosser, G, Ogrodowicz, R.W, de Carvalho, L.P.S. | | Deposit date: | 2022-10-04 | | Release date: | 2023-04-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure of the d-Cycloserine-Resistant Variant D322N of Alanine Racemase from Mycobacterium tuberculosis .
Acs Bio Med Chem Au, 3, 2023
|
|
6UXX
 
 | | PRMT5:MEP50 Complexed with Allosteric Inhibitor Compound 1a | | Descriptor: | (5R)-2-amino-5-(4-methoxyphenyl)-3-methyl-5-[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-yl]-3,5-dihydro-4H-imidazol-4-one, Methylosome protein 50, Protein arginine N-methyltransferase 5 | | Authors: | Palte, R.L, Schneider, S.E. | | Deposit date: | 2019-11-08 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Allosteric Modulation of Protein Arginine Methyltransferase 5 (PRMT5).
Acs Med.Chem.Lett., 11, 2020
|
|
6VEI
 
 | | Crystal Structure of Human Cytosolic Isocitrate Dehydrogenase (IDH1) R132H Mutant in Complex with NADPH and AG-881 (Vorasidenib) Inhibitor | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, 6-(6-chloropyridin-2-yl)-N2,N4-bis[(2R)-1,1,1-trifluoropropan-2-yl]-1,3,5-triazine-2,4-diamine, ACETATE ION, ... | | Authors: | Padyana, A, Jin, L. | | Deposit date: | 2020-01-02 | | Release date: | 2020-02-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Vorasidenib (AG-881): A First-in-Class, Brain-Penetrant Dual Inhibitor of Mutant IDH1 and 2 for Treatment of Glioma.
Acs Med.Chem.Lett., 11, 2020
|
|
6I83
 
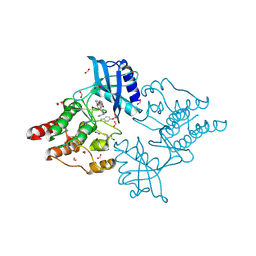 | | Crystal structure of phosphorylated RET V804M tyrosine kinase domain complexed with PDD00018366 | | Descriptor: | 4-[5-(pyridin-3-ylmethylamino)pyrazolo[1,5-a]pyrimidin-3-yl]benzamide, FORMIC ACID, Proto-oncogene tyrosine-protein kinase receptor Ret | | Authors: | Burschowsky, D, Seewooruthun, C, Bayliss, R, Carr, M.D, Echalier, A, Jordan, A.M. | | Deposit date: | 2018-11-19 | | Release date: | 2020-03-11 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Discovery and Optimization of wt-RET/KDR-Selective Inhibitors of RETV804MKinase.
Acs Med.Chem.Lett., 11, 2020
|
|
6WFN
 
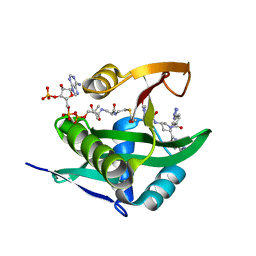 | | Crystal structure of human Naa50 in complex with AcCoA and an inhibitor (compound 4a) identified using DNA encoded library technology | | Descriptor: | (4S)-1-methyl-N-{(3S,5S)-5-[4-(methylcarbamoyl)-1,3-thiazol-2-yl]-1-[4-(1H-tetrazol-5-yl)benzene-1-carbonyl]pyrrolidin-3-yl}-2,6-dioxohexahydropyrimidine-4-carboxamide, ACETYL COENZYME *A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
6WF3
 
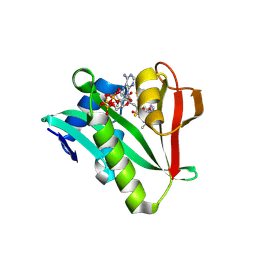 | | Crystal structure of human Naa50 in complex with a cofactor derived inhibitor (compound 1) | | Descriptor: | ACE-MET-LEU-GLY-PRO-NH2, COENZYME A, N-alpha-acetyltransferase 50 | | Authors: | Greasley, S.E, Feng, J, Deng, Y.-L, Stewart, A.E. | | Deposit date: | 2020-04-03 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Characterization of SpecificN-alpha-Acetyltransferase 50 (Naa50) Inhibitors Identified Using a DNA Encoded Library.
Acs Med.Chem.Lett., 11, 2020
|
|
7CBP
 
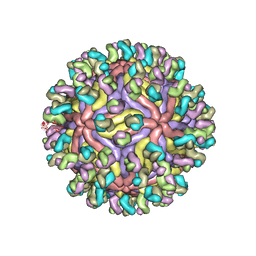 | | CryoEM structure of Zika virus with Fab at 4.1 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Fab Heavy chain, ... | | Authors: | Tyagi, A, Ahmed, T, Shi, J, Bhushan, S. | | Deposit date: | 2020-06-13 | | Release date: | 2020-07-08 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A complex between the Zika virion and the Fab of a broadly cross-reactive neutralizing monoclonal antibody revealed by cryo-EM and single particle analysis at 4.1 angstrom resolution.
J Struct Biol X, 4, 2020
|
|
5ZUN
 
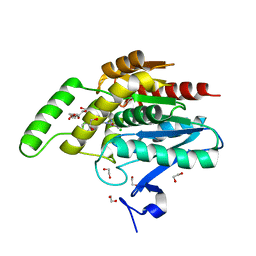 | | Crystal structure of human monoacylglycerol lipase in complex with compound 3l | | Descriptor: | (4R)-1-(2'-chloro[1,1'-biphenyl]-3-yl)-4-[4-(1,3-thiazole-2-carbonyl)piperazin-1-yl]pyrrolidin-2-one, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Sogabe, S, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2018-05-08 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Design, Synthesis, and Evaluation of Piperazinyl Pyrrolidin-2-ones as a Novel Series of Reversible Monoacylglycerol Lipase Inhibitors
J. Med. Chem., 61, 2018
|
|
5HG1
 
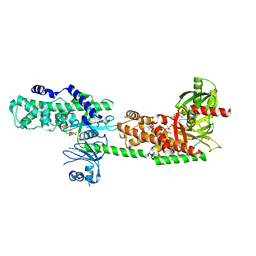 | | Crystal Structure of Human Hexokinase 2 with cmpd 1, a C-2-substituted glucosamine | | Descriptor: | 2-deoxy-2-{[(2E)-3-(3,4-dichlorophenyl)prop-2-enoyl]amino}-alpha-D-glucopyranose, 6-O-phosphono-beta-D-glucopyranose, CITRATE ANION, ... | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-07 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
5HEX
 
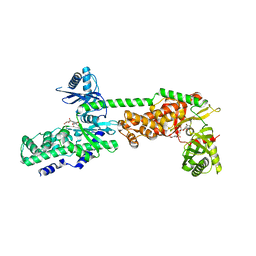 | | Crystal Structure of Human Hexokinase 2 with cmpd 30, a 2-amino-6-benzenesulfonamide glucosamine | | Descriptor: | 2-[(3-bromobenzene-1-carbonyl)amino]-6-{[(4-carboxy-5-methylfuran-2-yl)sulfonyl]amino}-2,6-dideoxy-alpha-D-glucopyranos e, Hexokinase-2 | | Authors: | Campobasso, N, Zhao, B, Smallwood, A. | | Deposit date: | 2016-01-06 | | Release date: | 2016-03-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Discovery of a Novel 2,6-Disubstituted Glucosamine Series of Potent and Selective Hexokinase 2 Inhibitors.
Acs Med.Chem.Lett., 7, 2016
|
|
7PFS
 
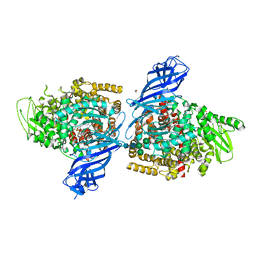 | | Crystal structure of ERAP2 aminopeptidase in complex with phosphinic pseudotripeptide ((1R)-1-Amino-3-phenylpropyl){2-([1,1:3,1-terphenyl]-5-ylmethyl)-3-[((2S)-1-amino-1-oxo-3-phenylpropan-2-yl)-amino]-3-oxopropyl}phosphinic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Giastas, P, Stratikos, E, Mpakali, A. | | Deposit date: | 2021-08-12 | | Release date: | 2022-01-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Inhibitor-Dependent Usage of the S1' Specificity Pocket of ER Aminopeptidase 2.
Acs Med.Chem.Lett., 13, 2022
|
|
8OKB
 
 | | SARS-CoV2 NSP5 in complex with a peptidomimetic ligand | | Descriptor: | 3C-like proteinase nsp5, methyl (4~{S})-4-[[(2~{S})-4-methyl-2-(phenylmethoxycarbonylamino)pentanoyl]amino]-5-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]pentanoate | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
8OKC
 
 | | SARS-CoV2 NSP5 in complex with a GC-376 based peptidomimetic PROTAC | | Descriptor: | (phenylmethyl) ~{N}-[(2~{R})-1-[[(~{Z},2~{S})-5-[4-[[1-[2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]-6-fluoranyl-1,3-bis(oxidanylidene)isoindol-5-yl]piperidin-4-yl]methyl]piperazin-1-yl]-5-oxidanylidene-1-[(3~{R})-2-oxidanylidenepyrrolidin-3-yl]pent-3-en-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate, 3C-like proteinase nsp5 | | Authors: | Calderone, V. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-17 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of a GC-376 Based Peptidomimetic PROTAC as a Degrader of 3-Chymotrypsin-like Protease of SARS-CoV-2.
Acs Med.Chem.Lett., 15, 2024
|
|
7B42
 
 | | Crystal structure of c-MET bound by compound 8 | | Descriptor: | 3-[(3-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3Q
 
 | | Crystal structure of c-MET bound by compound 1 | | Descriptor: | 1-(phenylmethyl)-5~{H}-pyrrolo[3,2-c]pyridin-4-one, Hepatocyte growth factor receptor, SULFATE ION | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B40
 
 | | Crystal structure of c-MET bound by compound 6 | | Descriptor: | 3-(phenylmethyl)-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3W
 
 | | Crystal structure of c-MET bound by compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 3-(6-fluoranyl-1~{H}-indazol-4-yl)-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B43
 
 | | Crystal structure of c-MET bound by compound 9 | | Descriptor: | 3-[(4-fluorophenyl)methyl]-5-(1-piperidin-4-ylpyrazol-4-yl)-1~{H}-pyrrolo[2,3-b]pyridine, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-02 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
7B3Z
 
 | | Crystal structure of c-MET bound by compound 5 | | Descriptor: | 3-[3-(phenylmethyl)-1~{H}-pyrrolo[2,3-b]pyridin-5-yl]-4,5-dihydro-1~{H}-pyrrolo[3,4-b]pyrrol-6-one, DIMETHYL SULFOXIDE, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2020-12-01 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis for Targeting the Folded P-Loop Conformation of c-MET.
Acs Med.Chem.Lett., 12, 2021
|
|
