1JQJ
 
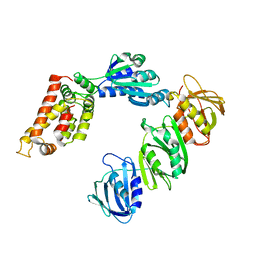 | | Mechanism of Processivity Clamp Opening by the Delta Subunit Wrench of the Clamp Loader Complex of E. coli DNA Polymerase III: Structure of the beta-delta complex | | Descriptor: | DNA polymerase III, beta chain, delta subunit | | Authors: | Jeruzalmi, D, Yurieva, O, Zhao, Y, Young, M, Stewart, J, Hingorani, M, O'Donnell, M, Kuriyan, J. | | Deposit date: | 2001-08-07 | | Release date: | 2001-11-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of processivity clamp opening by the delta subunit wrench of the clamp loader complex of E. coli DNA polymerase III.
Cell(Cambridge,Mass.), 106, 2001
|
|
1JYO
 
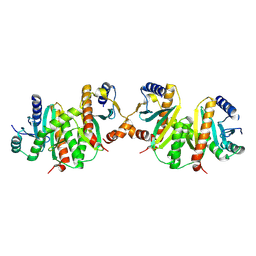 | |
2JNC
 
 | | Refined 3D NMR structure of ECD1 of mCRF-R2beta at pH 5 | | Descriptor: | Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JND
 
 | | 3D NMR structure of ECD1 of mCRF-R2b in complex with Astressin | | Descriptor: | ASTRESSIN, Corticotropin-releasing factor receptor 2 | | Authors: | Grace, C.R.R, Perrin, M.H, Jozsef, G, DiGruccio, M.R, Cantle, J.P, Rivier, J.E, Vale, W.W, Riek, R. | | Deposit date: | 2007-01-08 | | Release date: | 2007-03-13 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
2JOU
 
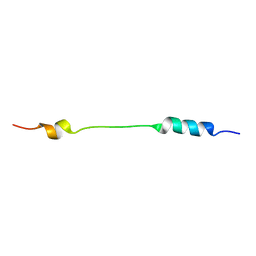 | | NMR structure of Mini-B, an N-terminal- C-terminal construct from human Surfactant Protein-B (SP-B), in Hexafluoroisopropanol (HFIP) | | Descriptor: | Pulmonary surfactant-associated protein B | | Authors: | Booth, V, Sarker, M, Keough, K.M.W, Waring, A.J, Walther, F.J. | | Deposit date: | 2007-03-26 | | Release date: | 2007-04-10 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structure of mini-B, a functional fragment of surfactant protein B, in detergent micelles
Biochemistry, 46, 2007
|
|
2J9V
 
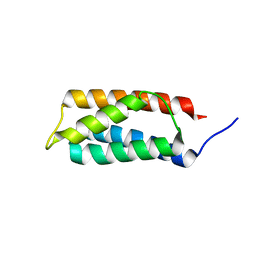 | | 2 Angstrom X-ray structure of the yeast ESCRT-I Vps28 C-terminus | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 28 | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
2J9U
 
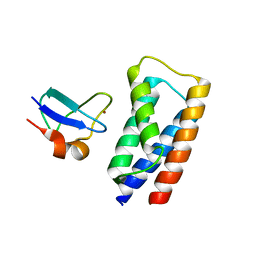 | | 2 Angstrom X-ray structure of the yeast ESCRT-I Vps28 C-terminus in complex with the NZF-N domain from ESCRT-II | | Descriptor: | VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 28, VACUOLAR PROTEIN SORTING-ASSOCIATED PROTEIN 36, ZINC ION | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
2J9W
 
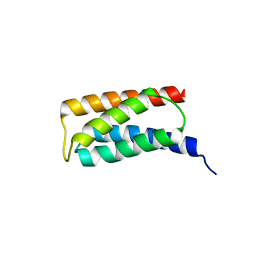 | | Structural insight into the ESCRT-I-II link and its role in MVB trafficking | | Descriptor: | VPS28-PROV PROTEIN | | Authors: | Gill, D.J, Teo, H.L, Sun, J, Perisic, O, Veprintsev, D.B, Emr, S.D, Williams, R.L. | | Deposit date: | 2006-11-16 | | Release date: | 2007-01-23 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Insight Into the Escrt-I/-II Link and its Role in Mvb Trafficking.
Embo J., 26, 2007
|
|
2JZH
 
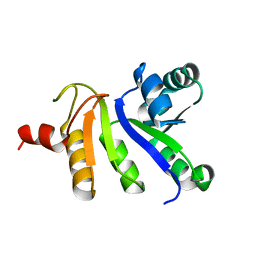 | | structure of IIB domain of the mannose transporter of E. coli | | Descriptor: | PTS system mannose-specific EIIAB component | | Authors: | Komlosh, M, Williams Jr, D.C. | | Deposit date: | 2008-01-08 | | Release date: | 2008-02-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structures of Productive and Non-productive Complexes between the A and B Domains of the Cytoplasmic Subunit of the Mannose Transporter of the Escherichia coli Phosphotransferase System.
J.Biol.Chem., 283, 2008
|
|
1M6E
 
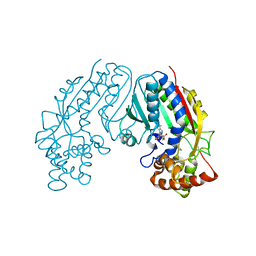 | | CRYSTAL STRUCTURE OF SALICYLIC ACID CARBOXYL METHYLTRANSFERASE (SAMT) | | Descriptor: | 2-HYDROXYBENZOIC ACID, LUTETIUM (III) ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zubieta, C, Ross, J.R, Koscheski, P, Yang, Y, Pichersky, E, Noel, J.P. | | Deposit date: | 2002-07-16 | | Release date: | 2003-09-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Basis for Substrate Recognition in The Salicylic Acid Carboxyl Methyltransferase Family
Plant Cell, 15, 2003
|
|
1M6S
 
 | | Crystal Structure Of Threonine Aldolase | | Descriptor: | CALCIUM ION, CHLORIDE ION, L-allo-threonine aldolase | | Authors: | Burley, S.K, Kielkopf, C.L. | | Deposit date: | 2002-07-17 | | Release date: | 2002-12-11 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray Structures of Threonine Aldolase Complexes: Structural Basis of Substrate Recognition
Biochemistry, 41, 2002
|
|
1MDY
 
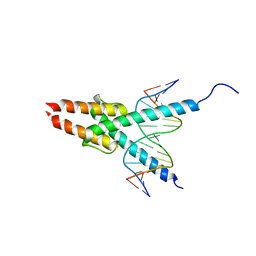 | | CRYSTAL STRUCTURE OF MYOD BHLH DOMAIN BOUND TO DNA: PERSPECTIVES ON DNA RECOGNITION AND IMPLICATIONS FOR TRANSCRIPTIONAL ACTIVATION | | Descriptor: | DNA (5'-D(*TP*CP*AP*AP*CP*AP*GP*CP*TP*GP*TP*TP*GP*A)-3'), PROTEIN (MYOD BHLH DOMAIN) | | Authors: | Ma, P.C.M, Rould, M.A, Weintraub, H, Pabo, C.O. | | Deposit date: | 1994-06-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation.
Cell(Cambridge,Mass.), 77, 1994
|
|
2JR5
 
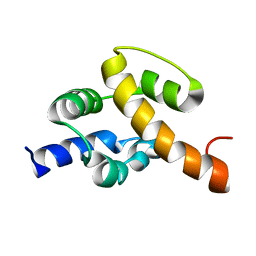 | | Solution structure of UPF0350 protein VC_2471. Northeast Structural Genomics Target VcR36 | | Descriptor: | UPF0350 protein VC_2471 | | Authors: | Wu, Y, Parish, D, Singarapu, K.K, Sukumaran, D, Eletski, A, Shastry, R, Nwosu, C, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of UPF0350 protein VC_2471.
To be Published
|
|
1MC3
 
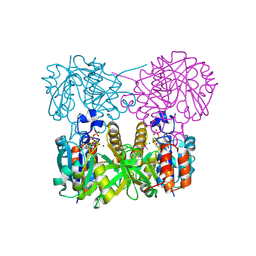 | | CRYSTAL STRUCTURE OF RFFH | | Descriptor: | GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE | | Authors: | Sivaraman, J, Sauve, V, Matte, A, Cygler, M, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2002-08-05 | | Release date: | 2002-11-20 | | Last modified: | 2018-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of Escherichia coli Glucose-1-Phosphate Thymidylyltransferase (RffH) Complexed with dTTP and Mg2+
J.BIOL.CHEM., 277, 2002
|
|
1MFM
 
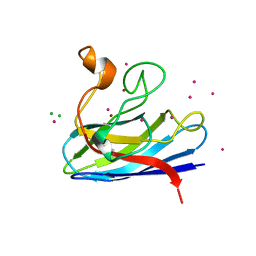 | | MONOMERIC HUMAN SOD MUTANT F50E/G51E/E133Q AT ATOMIC RESOLUTION | | Descriptor: | CADMIUM ION, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Ferraroni, M, Rypniewski, W, Wilson, K.S, Orioli, P.L, Viezzoli, M.S, Banci, L, Bertini, I, Mangani, S. | | Deposit date: | 1999-04-16 | | Release date: | 1999-04-21 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited.
J.Mol.Biol., 288, 1999
|
|
1ME9
 
 | |
1MEW
 
 | |
2LIS
 
 | |
2L7Z
 
 | | NMR Structure of A13 homedomain | | Descriptor: | Homeobox protein Hox-A13 | | Authors: | Ames, J. | | Deposit date: | 2010-12-27 | | Release date: | 2011-11-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis for sequence specific DNA binding and protein dimerization of HOXA13.
Plos One, 6, 2011
|
|
2MOI
 
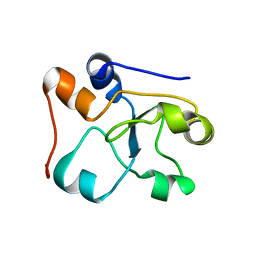 | | 3D NMR structure of the cytoplasmic rhodanese domain of the inner membrane protein YgaP from Escherichia coli | | Descriptor: | Inner membrane protein YgaP | | Authors: | Eichmann, C, Tzitzilonis, C, Bordignon, E, Maslennikov, I, Choe, S, Riek, R. | | Deposit date: | 2014-04-26 | | Release date: | 2014-06-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure and Functional Analysis of the Integral Membrane Protein YgaP from Escherichia coli.
J.Biol.Chem., 289, 2014
|
|
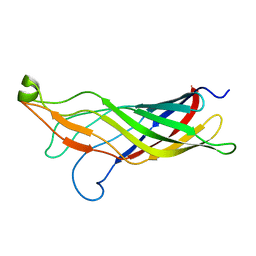
2MHL
 
 | |
2LWL
 
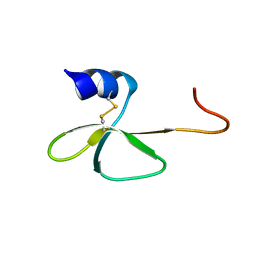 | | Structural Basis for the Interaction of Human β-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles | | Descriptor: | Beta-defensin 106 | | Authors: | de Paula, V.S, Gomes, N.S.F, Lima, L.G, Miyamoto, C.A, Monteiro, R.Q, Almeida, F.C.L, Valente, A. | | Deposit date: | 2012-08-02 | | Release date: | 2013-08-21 | | Last modified: | 2013-11-13 | | Method: | SOLUTION NMR | | Cite: | Structural Basis for the Interaction of Human beta-Defensin 6 and Its Putative Chemokine Receptor CCR2 and Breast Cancer Microvesicles.
J.Mol.Biol., 425, 2013
|
|
6E5X
 
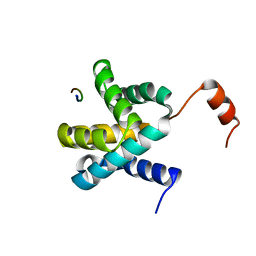 | | Crystal structure of Ebola virus VP30 C-terminus/RBBP6 peptide complex | | Descriptor: | CALCIUM ION, E3 ubiquitin-protein ligase RBBP6, Minor nucleoprotein VP30 | | Authors: | Liu, D, Small, G.I, Leung, D.W, Amarasinghe, G.K. | | Deposit date: | 2018-07-23 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Protein Interaction Mapping Identifies RBBP6 as a Negative Regulator of Ebola Virus Replication.
Cell, 175, 2018
|
|
6I3M
 
 | | eIF2B:eIF2 complex, phosphorylated on eIF2 alpha serine 52. | | Descriptor: | Eukaryotic translation initiation factor 2 subunit alpha, Eukaryotic translation initiation factor 2 subunit beta, Eukaryotic translation initiation factor 2 subunit gamma, ... | | Authors: | Adomavicius, T, Roseman, A.M, Pavitt, G.D. | | Deposit date: | 2018-11-06 | | Release date: | 2019-05-22 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | The structural basis of translational control by eIF2 phosphorylation.
Nat Commun, 10, 2019
|
|
7JZV
 
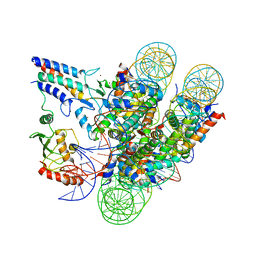 | | Cryo-EM structure of the BRCA1-UbcH5c/BARD1 E3-E2 module bound to a nucleosome | | Descriptor: | BRCA1,Ubiquitin-conjugating enzyme E2 D3, BRCA1-associated RING domain protein 1, Histone H2A type 2-A, ... | | Authors: | Witus, S.R, Burrell, A.L, Hansen, J.M, Farrell, D.P, Dimaio, F, Kollman, J.M, Klevit, R.E. | | Deposit date: | 2020-09-02 | | Release date: | 2021-02-17 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | BRCA1/BARD1 site-specific ubiquitylation of nucleosomal H2A is directed by BARD1.
Nat.Struct.Mol.Biol., 28, 2021
|
|
