5A9U
 
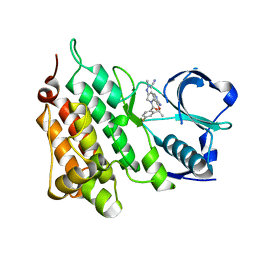 | | Structure of C1156Y Mutant Human Anaplastic Lymphoma Kinase in Complex with PF-06463922 ((10R)-7-amino-12-fluoro-2,10,16-trimethyl- 15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo(4,3-h)(2,5,11) benzoxadiazacyclotetradecine-3-carbonitrile). | | Descriptor: | (10R)-7-amino-12-fluoro-2,10,16-trimethyl-15-oxo-10,15,16,17-tetrahydro-2H-8,4-(metheno)pyrazolo[4,3-h][2,5,11]benzoxadiazacyclotetradecine-3-carbonitrile, ALK TYROSINE KINASE RECEPTOR | | Authors: | McTigue, M, Deng, Y.-L, Liu, W, Brooun, A, Stewart, A. | | Deposit date: | 2015-07-22 | | Release date: | 2016-06-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Resensitization to Crizotinib by the Lorlatinib Alk Resistance Mutation L1198F.
N.Engl.J.Med., 374, 2016
|
|
5FA6
 
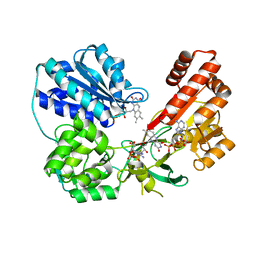 | | wild type human CYPOR | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | Deposit date: | 2015-12-11 | | Release date: | 2016-08-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
5H59
 
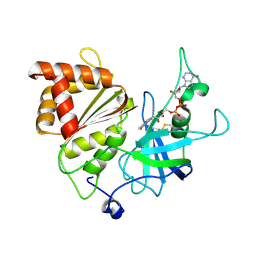 | | Ferredoxin-NADP+ reductase from maize root | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Kurisu, G, Hase, T. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the isotype-specific interactions of ferredoxin and ferredoxin: NADP(+) oxidoreductase: an evolutionary switch between photosynthetic and heterotrophic assimilation
Photosyn. Res., 134, 2017
|
|
5UG8
 
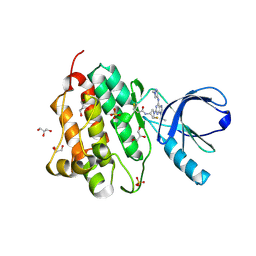 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | Epidermal growth factor receptor, GLYCEROL, N-[(3R,4R)-4-fluoro-1-{6-[(1-methyl-1H-pyrazol-4-yl)amino]-9-(propan-2-yl)-9H-purin-2-yl}pyrrolidin-3-yl]propanamide, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-07 | | Release date: | 2017-03-22 | | Last modified: | 2017-04-26 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5H28
 
 | |
5GRT
 
 | | HUMAN GLUTATHIONE REDUCTASE A34E, R37W MUTANT, GLUTATHIONYLSPERMIDINE COMPLEX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLUTATHIONE REDUCTASE, GLUTATHIONYLSPERMIDINE DISULFIDE | | Authors: | Stoll, V.S, Simpson, S.J, Krauth-Siegel, R.L, Walsh, C.T, Pai, E.F. | | Deposit date: | 1997-02-12 | | Release date: | 1997-08-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Glutathione reductase turned into trypanothione reductase: structural analysis of an engineered change in substrate specificity.
Biochemistry, 36, 1997
|
|
2OKX
 
 | | Crystal structure of GH78 family rhamnosidase of Bacillus SP. GL1 AT 1.9 A | | Descriptor: | CALCIUM ION, GLYCEROL, Rhamnosidase B | | Authors: | Cui, Z, Mikami, B, Hashimoto, W, Murata, K. | | Deposit date: | 2007-01-17 | | Release date: | 2007-11-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Glycoside Hydrolase Family 78 alpha-L-Rhamnosidase from Bacillus sp. GL1
J.Mol.Biol., 374, 2007
|
|
5URC
 
 | | Design, Synthesis, Functional and Biological Evaluation of Ether and Ester Derivatives of the Antisickling Agent 5-HMF for the Treatment of Sickle Cell Disease | | Descriptor: | (5-formylfuran-2-yl)methyl acetate, 5-HYDROXYMETHYL-FURFURAL, CARBON MONOXIDE, ... | | Authors: | Pagare, P.P, Safo, R.S, Gazi, A. | | Deposit date: | 2017-02-10 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Design, Synthesis, and Biological Evaluation of Ester and Ether Derivatives of Antisickling Agent 5-HMF for the Treatment of Sickle Cell Disease.
Mol. Pharm., 14, 2017
|
|
5CXM
 
 | | Crystal structure of the cyanobacterial plasma membrane Rieske protein PetC3 from Synechocystis PCC 6803 | | Descriptor: | Cytochrome b6/f complex iron-sulfur subunit, FE2/S2 (INORGANIC) CLUSTER, NICKEL (II) ION, ... | | Authors: | Veit, S, Takeda, K, Miki, K, Roegner, M. | | Deposit date: | 2015-07-29 | | Release date: | 2016-08-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and functional characterisation of the cyanobacterial PetC3 Rieske protein family.
Biochim. Biophys. Acta, 1857, 2016
|
|
5EMN
 
 | | Crystal Structure of Human NADPH-Cytochrome P450 Reductase(A287P mutant) | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Xia, C, Marohnic, C, Panda, S, Masters, B.S, Kim, J.J.K. | | Deposit date: | 2015-11-06 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Instability of the Human Cytochrome P450 Reductase A287P Variant Is the Major Contributor to Its Antley-Bixler Syndrome-like Phenotype.
J.Biol.Chem., 291, 2016
|
|
5GQD
 
 | | Crystal structure of covalent glycosyl-enzyme intermediate of xylanase mutant (T82A, N127S, and E128H) from Streptomyces olivaceoviridis E-86 | | Descriptor: | Beta-xylanase, GLYCEROL, beta-D-xylopyranose-(1-4)-alpha-D-xylopyranose | | Authors: | Suzuki, R, Fujimoto, Z, Kaneko, S, Kuno, A. | | Deposit date: | 2016-08-07 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Azidolysis by the Formation of Stable Ser-His Catalytic Dyad in a Glycoside Hydrolase Family 10 Xylanase Mutant
J.Appl.Glyosci., 65, 2019
|
|
5DAA
 
 | |
5H1H
 
 | | NMR structure of SLBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
5H1I
 
 | | NMR structure of TIBA, a chimera of SFTI | | Descriptor: | Bradykinin-trypsin inhibitor secondary loop chimera | | Authors: | Xiao, T, Tam, J.P. | | Deposit date: | 2016-10-10 | | Release date: | 2017-04-19 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | An Orally Active Bradykinin B1 Receptor Antagonist Engineered as a Bifunctional Chimera of Sunflower Trypsin Inhibitor.
J. Med. Chem., 60, 2017
|
|
5E9G
 
 | | Structural insights of isocitrate lyases from Magnaporthe oryzae | | Descriptor: | GLYCEROL, GLYOXYLIC ACID, Isocitrate lyase, ... | | Authors: | Park, Y, Cho, Y, Lee, Y.-H, Lee, Y.-W, Rhee, S. | | Deposit date: | 2015-10-15 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure and functional analysis of isocitrate lyases from Magnaporthe oryzae and Fusarium graminearum
J.Struct.Biol., 194, 2016
|
|
5AVP
 
 | |
5AY2
 
 | | Crystal structure of RNA duplex containing C-Ag(I)-C base pairs | | Descriptor: | RNA (5'-R(*GP*GP*AP*CP*UP*(CBR)P*GP*AP*CP*UP*CP*C)-3'), SILVER ION | | Authors: | Kondo, J, Tada, Y, Dairaku, T, Saneyoshi, H, Okamoto, I, Tanaka, Y, Ono, A. | | Deposit date: | 2015-08-06 | | Release date: | 2015-10-21 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Crystal Structure of a Silver(I)-RNA Hybrid Duplex Containing Watson-Crick-like CSilver(I)C Metallo-Base Pairs
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5EAA
 
 | | ASPARTATE AMINOTRANSFERASE FROM E. COLI, C191S MUTATION | | Descriptor: | ASPARTATE AMINOTRANSFERASE, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Jeffery, C.J, Gloss, L.M, Petsko, G.A, Ringe, D. | | Deposit date: | 1998-12-29 | | Release date: | 2000-10-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The role of residues outside the active site: structural basis for function of C191 mutants of Escherichia coli aspartate aminotransferase.
Protein Eng., 13, 2000
|
|
5EAT
 
 | | 5-EPI-ARISTOLOCHENE SYNTHASE FROM NICOTIANA TABACUM WITH SUBSTRATE ANALOG FARNESYL HYDROXYPHOSPHONATE | | Descriptor: | 1-HYDROXY-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENE PHOSPHONIC ACID, 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION | | Authors: | Starks, C.M, Back, K, Chappell, J, Noel, J.P. | | Deposit date: | 1997-07-24 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase.
Science, 277, 1997
|
|
5EAS
 
 | | 5-EPI-ARISTOLOCHENE SYNTHASE FROM NICOTIANA TABACUM | | Descriptor: | 5-EPI-ARISTOLOCHENE SYNTHASE, MAGNESIUM ION | | Authors: | Starks, C.M, Back, K, Chappell, J, Noel, J.P. | | Deposit date: | 1997-06-19 | | Release date: | 1997-10-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase.
Science, 277, 1997
|
|
5ER2
 
 | | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor. the analysis of the inhibitor binding and description of the rigid body shift in the enzyme | | Descriptor: | 6-ammonio-N-{[(2R,3R)-3-{[N-(tert-butoxycarbonyl)-L-phenylalanyl-3-(1H-imidazol-3-ium-4-yl)-L-alanyl]amino}-4-cyclohexyl-2-hydroxybutyl](2-methylpropyl)carbamoyl}-L-norleucyl-L-phenylalanine, ENDOTHIAPEPSIN | | Authors: | Sali, A, Veerapandian, B, Cooper, J.B, Foundling, S.I, Hoover, D.J, Blundell, T.L. | | Deposit date: | 1991-01-02 | | Release date: | 1991-04-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-resolution X-ray diffraction study of the complex between endothiapepsin and an oligopeptide inhibitor: the analysis of the inhibitor binding and description of the rigid body shift in the enzyme.
EMBO J., 8, 1989
|
|
5ER1
 
 | |
4B3S
 
 | | Crystal structure of the 30S ribosome in complex with compound 37 | | Descriptor: | (1R,2R,3S,4R,6S)-4,6-diamino-2-{[3-O-(2,6-diamino-2,6-dideoxy-beta-L-idopyranosyl)-beta-D-ribofuranosyl]oxy}-3-hydroxycyclohexyl 2-amino-4-O-benzyl-2-deoxy-alpha-D-glucopyranoside, 16S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Ng, C.L, Lang, K, Shcherbakov, D, Matt, T, Perez-Fernandez, D, Patak, R, Meyer, M, Duscha, S, Akbergenov, R, Boukari, H, Freihofer, P, Kudyba, I, Reddy, M.S.K, Nandurikar, R.S, Ramakrishnan, V, Vasella, A, Bottger, E.C. | | Deposit date: | 2012-07-26 | | Release date: | 2013-08-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | 4'-O-Substitutions Determine Selectivity of Aminoglycoside Antibiotics
Nat.Commun., 5, 2014
|
|
5UGC
 
 | | Crystal structure of the EGFR kinase domain (L858R, T790M, V948R) in complex with a covalent inhibitor N-[(3R,4R)-4-fluoro-1-{6-[(3-methoxy-1-methyl-1H-pyrazol-4-yl)amino]-9-methyl-9H-purin-2-yl}pyrrolidin-3-yl]propanamide | | Descriptor: | 1,2-ETHANEDIOL, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S, Ferre, R.A. | | Deposit date: | 2017-01-08 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of N-((3R,4R)-4-Fluoro-1-(6-((3-methoxy-1-methyl-1H-pyrazol-4-yl)amino)-9-methyl-9H-purin-2-yl)pyrrolidine-3-yl)acrylamide (PF-06747775) through Structure-Based Drug Design: A High Affinity Irreversible Inhibitor Targeting Oncogenic EGFR Mutants with Selectivity over Wild-Type EGFR.
J. Med. Chem., 60, 2017
|
|
5HG9
 
 | | EGFR (L858R, T790M, V948R) in complex with 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]prop-2-en-1-one | | Descriptor: | 1-[(3R,4R)-3-[({2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-(trifluoromethyl)pyrrolidin-1-yl]propan-1-one, Epidermal growth factor receptor, GLYCEROL, ... | | Authors: | Gajiwala, K.S. | | Deposit date: | 2016-01-08 | | Release date: | 2016-02-03 | | Last modified: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of 1-{(3R,4R)-3-[({5-Chloro-2-[(1-methyl-1H-pyrazol-4-yl)amino]-7H-pyrrolo[2,3-d]pyrimidin-4-yl}oxy)methyl]-4-methoxypyrrolidin-1-yl}prop-2-en-1-one (PF-06459988), a Potent, WT Sparing, Irreversible Inhibitor of T790M-Containing EGFR Mutants.
J.Med.Chem., 59, 2016
|
|
