1HIB
 
 | | THE STRUCTURE OF AN INTERLEUKIN-1 BETA MUTANT WITH REDUCED BIOACTIVITY SHOWS MULTIPLE SUBTLE CHANGES IN CONFORMATION THAT AFFECT PROTEIN-PROTEIN RECOGNITION | | Descriptor: | INTERLEUKIN-1 BETA | | Authors: | Camacho, N.P, Smith, D.R, Goldman, A, Schneider, B, Green, D, Young, P.R, Berman, H.M. | | Deposit date: | 1993-03-29 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of an interleukin-1 beta mutant with reduced bioactivity shows multiple subtle changes in conformation that affect protein-protein recognition.
Biochemistry, 32, 1993
|
|
4BK7
 
 | |
5PZN
 
 | |
1HQ0
 
 | | CRYSTAL STRUCTURE OF THE CATALYTIC DOMAIN OF E.COLI CYTOTOXIC NECROTIZING FACTOR TYPE 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2000-12-13 | | Release date: | 2001-07-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
7WH7
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAF1_N179S) with xylotetraose | | Descriptor: | Beta-xylanase, alpha-D-mannopyranose-(1-2)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, G.Q, Zhang, R.F. | | Deposit date: | 2021-12-30 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | The mutant crystal structure of b-1,4-Xylanase (XynAF1_N179S) with xylotetraose
To Be Published
|
|
4ECM
 
 | | 2.3 Angstrom Crystal Structure of a Glucose-1-phosphate Thymidylyltransferase from Bacillus anthracis in Complex with Thymidine-5-diphospho-alpha-D-glucose and Pyrophosphate | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase, PYROPHOSPHATE 2- | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4BKD
 
 | |
3P5L
 
 | |
1TOZ
 
 | | NMR structure of the human NOTCH-1 ligand binding region | | Descriptor: | Neurogenic locus notch homolog protein 1 | | Authors: | Hambleton, S, Valeyev, N.Y, Muranyi, A, Knott, V, Werner, J.M, Mcmichael, A.J, Handford, P.A, Downing, A.K. | | Deposit date: | 2004-06-15 | | Release date: | 2004-10-12 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural and functional properties of the human notch-1 ligand binding region
STRUCTURE, 12, 2004
|
|
1PXE
 
 | |
1TR1
 
 | | CRYSTAL STRUCTURE OF E96K MUTATED BETA-GLUCOSIDASE A FROM BACILLUS POLYMYXA, AN ENZYME WITH INCREASED THERMORESISTANCE | | Descriptor: | BETA-GLUCOSIDASE A, GLYCEROL | | Authors: | Sanz-Aparicio, J, Hermoso, J.A, Martinez-Ripoll, M, Gonzalez-Perez, B, Polaina, J. | | Deposit date: | 1998-03-12 | | Release date: | 1999-04-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of beta-glucosidase A from Bacillus polymyxa: insights into the catalytic activity in family 1 glycosyl hydrolases.
J.Mol.Biol., 275, 1998
|
|
1HZG
 
 | | CRYSTAL STRUCTURE OF THE INACTIVE C866S MUTANT OF THE CATALYTIC DOMAIN OF E. COLI CYTOTOXIC NECROTIZING FACTOR 1 | | Descriptor: | CYTOTOXIC NECROTIZING FACTOR 1, PHOSPHATE ION | | Authors: | Buetow, L, Flatau, G, Chiu, K, Boquet, P, Ghosh, P. | | Deposit date: | 2001-01-24 | | Release date: | 2001-07-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of the Rho-activating domain of Escherichia coli cytotoxic necrotizing factor 1.
Nat.Struct.Biol., 8, 2001
|
|
2M8M
 
 | | NMR assignment and structure of a peptide derived from the membrane proximal external region of HIV-1 gp41 in the presence of hexafluoroisopropanol | | Descriptor: | Transmembrane protein gp41 | | Authors: | Serrano, S, Huarte, N, Nieva, J.L, Jimenez, M. | | Deposit date: | 2013-05-23 | | Release date: | 2014-01-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Immunogenicity of a Peptide Vaccine, Including the Complete HIV-1 gp41 2F5 Epitope: IMPLICATIONS FOR ANTIBODY RECOGNITION MECHANISM AND IMMUNOGEN DESIGN.
J.Biol.Chem., 289, 2014
|
|
1XI7
 
 | | NMR structure of the carboxyl-terminal cysteine domain of the VHv1.1 polydnaviral gene product | | Descriptor: | cysteine-rich omega-conotoxin homolog VHv1.1 | | Authors: | Einerwold, J, Jaseja, M, Hapner, K, Webb, B, Copie, V. | | Deposit date: | 2004-09-21 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the carboxyl-terminal cysteine-rich domain of the VHv1.1 polydnaviral gene product: comparison with other cystine knot structural folds
Biochemistry, 40, 2001
|
|
4EXP
 
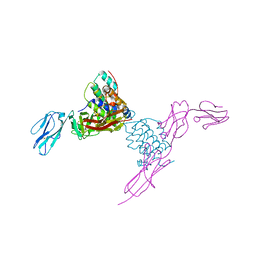 | | Structure of mouse Interleukin-34 in complex with mouse FMS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Interleukin-34, Macrophage colony-stimulating factor 1 receptor | | Authors: | Liu, H, Leo, C, Chen, X, Wong, B.R, Williams, L.T, Lin, H, He, X. | | Deposit date: | 2012-04-30 | | Release date: | 2012-05-30 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The mechanism of shared but distinct CSF-1R signaling by the non-homologous cytokines IL-34 and CSF-1.
Biochim.Biophys.Acta, 1824, 2012
|
|
1XBT
 
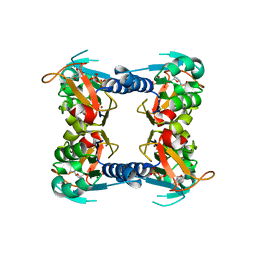 | | Crystal Structure of Human Thymidine Kinase 1 | | Descriptor: | MAGNESIUM ION, THYMIDINE-5'-TRIPHOSPHATE, Thymidine kinase, ... | | Authors: | Welin, M, Kosinska, U, Mikkelsen, N.E, Carnrot, C, Zhu, C, Wang, L, Eriksson, S, Munch-Petersen, B, Eklund, H. | | Deposit date: | 2004-08-31 | | Release date: | 2004-12-14 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of thymidine kinase 1 of human and mycoplasmic origin
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1TS5
 
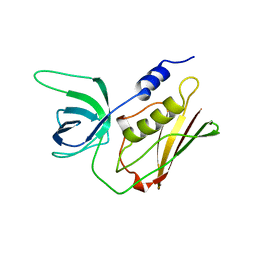 | | I140T MUTANT OF TOXIC SHOCK SYNDROME TOXIN-1 FROM S. AUREUS | | Descriptor: | TOXIC SHOCK SYNDROME TOXIN-1 | | Authors: | Earhart, C.A, Mitchell, D.T, Murray, D.L, Pinheiro, D.M, Matsumura, M, Schlievert, P.M, Ohlendorf, D.H. | | Deposit date: | 1997-10-10 | | Release date: | 1998-12-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of five mutants of toxic shock syndrome toxin-1 with reduced biological activity.
Biochemistry, 37, 1998
|
|
2RIJ
 
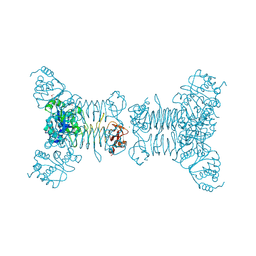 | | Crystal structure of a putative 2,3,4,5-tetrahydropyridine-2-carboxylate n-succinyltransferase (cj1605c, dapd) from campylobacter jejuni at 1.90 A resolution | | Descriptor: | CHLORIDE ION, CITRIC ACID, GLYCEROL, ... | | Authors: | Joint Center for Structural Genomics (JCSG) | | Deposit date: | 2007-10-11 | | Release date: | 2007-10-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Putative 2,3,4,5-tetrahydropyridine-2-carboxylate N-succinyltransferase (NP_282733.1) from Campylobacter jejuni at 1.90 A resolution
To be published
|
|
1I7B
 
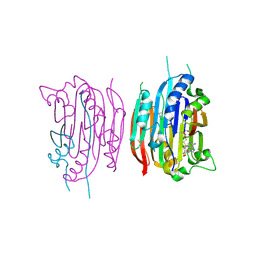 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND S-ADENOSYLMETHIONINE METHYL ESTER | | Descriptor: | 1,4-DIAMINOBUTANE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, S-ADENOSYLMETHIONINE DECARBOXYLASE BETA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
4E8C
 
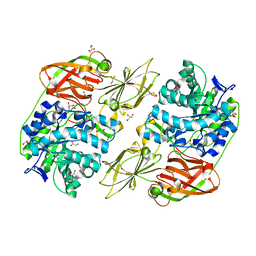 | | Crystal structure of streptococcal beta-galactosidase in complex with galactose | | Descriptor: | GLYCEROL, Glycosyl hydrolase, family 35, ... | | Authors: | Cheng, W, Wang, L, Bai, X.H, Jiang, Y.L, Li, Q, Yu, G, Zhou, C.Z, Chen, Y.X. | | Deposit date: | 2012-03-20 | | Release date: | 2012-05-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural insights into the substrate specificity of Streptococcus pneumoniae beta (1,3)-galactosidase BgaC
J.Biol.Chem., 287, 2012
|
|
6T9P
 
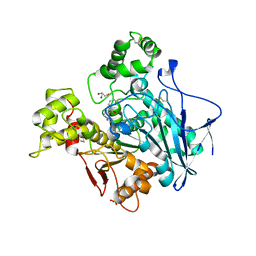 | | Human Butyrylcholinesterase in complex with 2-(N-hydroxyimino)-N-[(1R)-3-{4-[(2-methyl-1H-imidazol-1-yl)methyl]-1H-1,2,3-triazol-1-yl}-1- phenylpropyl]acetamide | | Descriptor: | (R,E)-2-(hydroxyimino)-N-(3-(4-((2-methyl-1H-imidazol-1-yl)methyl)-1H-1,2,3-triazol-1-yl)-1-phenylpropyl)acetamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[1-deoxy-alpha-D-tagatopyranose-(2-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Brazzolotto, X, Sinko, G, Marakovic, N, Knezevic, A. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Enantioseparation, in vitro testing, and structural characterization of triple-binding reactivators of organophosphate-inhibited cholinesterases.
Biochem.J., 477, 2020
|
|
3OSE
 
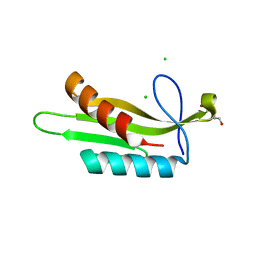 | |
4EVF
 
 | |
4OR5
 
 | | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4 | | Descriptor: | AF4/FMR2 family member 4, Cyclin-T1, Cyclin-dependent kinase 9, ... | | Authors: | Gu, J, Babayeva, N.D, Suwa, Y, Baranovskiy, A.G, Price, D.H, Tahirov, T.H. | | Deposit date: | 2014-02-10 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of HIV-1 Tat complexed with human P-TEFb and AFF4.
Cell Cycle, 13, 2014
|
|
3LQE
 
 | |
