6MRV
 
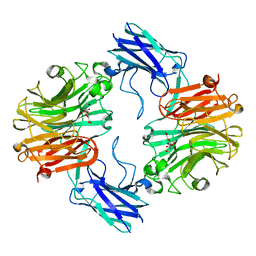 | | Sialidase26 co-crystallized with DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, Sialidase26 | | Authors: | Zaramela, L.S, Martino, C, Alisson-Silva, F, Rees, S.D, Diaz, S.L, Chuzel, L, Ganatra, M.B, Taron, C.H, Zuniga, C, Chang, G, Varki, A, Zengler, K. | | Deposit date: | 2018-10-15 | | Release date: | 2019-10-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Gut bacteria responding to dietary change encode sialidases that exhibit preference for red meat-associated carbohydrates.
Nat Microbiol, 4, 2019
|
|
5U5Y
 
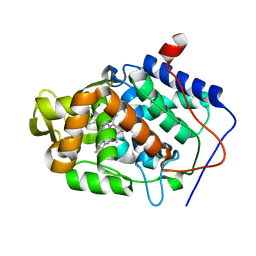 | | CcP gateless cavity | | Descriptor: | 2-methylimidazo[1,2-a]pyridin-3-amine, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8KB8
 
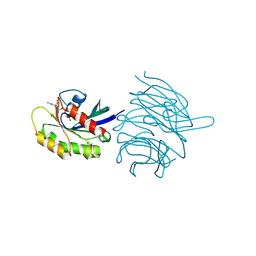 | | Structure of the WDR91 WD40 domain complexed with Rab7 | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Ras-related protein Rab-7a, ... | | Authors: | Li, J, Ma, X.L, Banerjee, S, Dong, Z.G. | | Deposit date: | 2023-08-04 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Insights into the distinct membrane targeting mechanisms of WDR91 family proteins.
Structure, 2024
|
|
8SBV
 
 | |
5H4I
 
 | |
7P32
 
 | | Crystal structure of human lysosomal acid-alpha-glucosidase, GAA, in complex with cyclosulfamidate 6 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Roig-Zamboni, V, Kok, K, Overkleeft, H, Artola, M, Sulzenbacher, G. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-13 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | 1,6- epi-Cyclophellitol Cyclosulfamidate Is a Bona Fide Lysosomal alpha-Glucosidase Stabilizer for the Treatment of Pompe Disease.
J.Am.Chem.Soc., 144, 2022
|
|
5TPH
 
 | | Crystal structure of a de novo designed protein homodimer with curved beta-sheet | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, de novo NTF2 homodimer | | Authors: | Basanta, B, Marcos, E, Oberdorfer, G, Chidyausiku, T.M, Sankaran, B, Baker, D. | | Deposit date: | 2016-10-20 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Principles for designing proteins with cavities formed by curved beta sheets.
Science, 355, 2017
|
|
8KAO
 
 | | Glutamate dehydrogenase-69O | | Descriptor: | 2-oxopentanoic acid, Glutamate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sakuraba, H, Ohshima, T. | | Deposit date: | 2023-08-03 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of glutamate dehydrogenase-69O
To be published
|
|
8SBO
 
 | |
5U60
 
 | | CcP gateless cavity | | Descriptor: | (4E)-4-hydrazinylidene-1,4-dihydroquinoline, PROTOPORPHYRIN IX CONTAINING FE, Peroxidase | | Authors: | Fischer, M, Shoichet, B.K. | | Deposit date: | 2016-12-07 | | Release date: | 2017-02-01 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Testing inhomogeneous solvation theory in structure-based ligand discovery.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8SAC
 
 | |
5CKL
 
 | |
5U72
 
 | |
8KD8
 
 | |
5CKY
 
 | | Crystal Structure of the MTERF1 R162A substitution bound to the termination sequence. | | Descriptor: | 5' -D (*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3', 5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3', Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-15 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
8V7R
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132 | | Descriptor: | (5R)-5-[2-(4-methoxyphenyl)ethyl]-5-methylimidazolidine-2,4-dione, 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-12-04 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z56772132
To Be Published
|
|
7P33
 
 | | Epstein-Barr virus encoded Bcl-2 homolog BHRF-1 in complex with Bid BH3 peptide | | Descriptor: | 1,2-ETHANEDIOL, Apoptosis regulator BHRF1, BH3-interacting domain death agonist p15, ... | | Authors: | Suraweera, C.D, Hinds, M.G, Kvansakul, M. | | Deposit date: | 2021-07-07 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.78542733 Å) | | Cite: | Crystal Structures of Epstein-Barr Virus Bcl-2 Homolog BHRF1 Bound to Bid and Puma BH3 Motif Peptides.
Viruses, 14, 2022
|
|
8KDU
 
 | | Crystal structure of trypsin with its substrate | | Descriptor: | (2~{R})-2-benzamido-5-carbamimidamido-pentanoic acid, ARGININE, CALCIUM ION, ... | | Authors: | Akbar, Z, Ahmad, M.S. | | Deposit date: | 2023-08-10 | | Release date: | 2024-08-14 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Michaelis complex of trypsin with N-alpha-benzoyl-l-arginine ethyl ester
J Chin Chem Soc, n/a, 2024
|
|
7Y0L
 
 | | SulE-S209A | | Descriptor: | Alpha/beta fold hydrolase, GLYCEROL, METHYL 2-[({[(4-METHOXY-6-METHYL-1,3,5-TRIAZIN-2-YL)AMINO]CARBONYL}AMINO)SULFONYL]BENZOATE | | Authors: | Liu, B, Ran, T, He, J, Wang, W. | | Deposit date: | 2022-06-05 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Crystal structures of herbicide-detoxifying esterase reveal a lid loop affecting substrate binding and activity.
Nat Commun, 14, 2023
|
|
8SBW
 
 | | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, orthorhombic form) | | Descriptor: | 2,3-dihydroxybenzoate-2,3-dehydrogenase, ACETATE ION, SULFATE ION | | Authors: | Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2023-04-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of 2,3-dihydro-2,3-dihydroxybenzoate dehydrogenase from Klebsiella aerogenes (Apo, orthorhombic form)
To be published
|
|
8KC4
 
 | | De novo design protein -NA05 | | Descriptor: | De novo design protein -NA05 | | Authors: | Wang, S, Liu, Y. | | Deposit date: | 2023-08-06 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | De novo design protein -NA05
To Be Published
|
|
5C0M
 
 | | Crystal structure of SGF29 tandem tudor domain in complex with a Carba containing peptide | | Descriptor: | Carba-containing peptide, GLYCEROL, SAGA-associated factor 29 homolog, ... | | Authors: | Dong, A, Xu, C, Tempel, W, Cerovina, T, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-06-12 | | Release date: | 2015-07-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Chemical basis for the recognition of trimethyllysine by epigenetic reader proteins.
Nat Commun, 6, 2015
|
|
7P2W
 
 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
8JY1
 
 | |
7P2X
 
 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | Descriptor: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | Authors: | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | Deposit date: | 2021-07-06 | | Release date: | 2022-07-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
