3GQL
 
 | | Crystal Structure of activated receptor tyrosine kinase in complex with substrates | | Descriptor: | (E)-[4-(3,5-difluorophenyl)-3H-pyrrolo[2,3-b]pyridin-3-ylidene](3-methoxyphenyl)methanol, Basic fibroblast growth factor receptor 1 | | Authors: | Bae, J.H, Lew, E.D, Yuzawa, S, Tome, F, Lax, I, Schlessinger, J. | | Deposit date: | 2009-03-24 | | Release date: | 2009-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The selectivity of receptor tyrosine kinase signaling is controlled by a secondary SH2 domain binding site.
Cell(Cambridge,Mass.), 138, 2009
|
|
2NYM
 
 | | Crystal Structure of Protein Phosphatase 2A (PP2A) with C-terminus truncated catalytic subunit | | Descriptor: | MANGANESE (II) ION, Protein phosphatase 2, Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit gamma isoform, ... | | Authors: | Chen, Y, Xing, Y, Xu, Y, Chao, Y, Lin, Z, Jeffrey, P.D, Shi, Y. | | Deposit date: | 2006-11-21 | | Release date: | 2006-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure of the Protein Phosphatase 2A Holoenzyme.
Cell(Cambridge,Mass.), 127, 2006
|
|
3SPU
 
 | | apo NDM-1 Crystal Structure | | Descriptor: | Beta-lactamase NDM-1, ZINC ION | | Authors: | Strynadka, N.C.J, King, D.T. | | Deposit date: | 2011-07-03 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of New Delhi metallo-beta-lactamase reveals molecular basis for antibiotic resistance
Protein Sci., 20, 2011
|
|
3ST8
 
 | |
3SUE
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-dioxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadecino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3GW0
 
 | | UROD mutant G318R | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3SQN
 
 | | Putative Mga family transcriptional regulator from Enterococcus faecalis | | Descriptor: | Conserved domain protein | | Authors: | Osipiuk, J, Wu, R, Jedrzejczak, R, Moy, S, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-07-05 | | Release date: | 2011-07-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Putative Mga family transcriptional regulator from Enterococcus faecalis.
To be Published
|
|
2NY1
 
 | | HIV-1 gp120 Envelope Glycoprotein (I109C, T257S, S334A, S375W, Q428C) Complexed with CD4 and Antibody 17b | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, HEAVY CHAIN, ... | | Authors: | Zhou, T, Xu, L, Dey, B, Hessell, A.J, Van Ryk, D, Xiang, S.H, Yang, X, Zhang, M.Y, Zwick, M.B, Arthos, J, Burton, D.R, Dimitrov, D.S, Sodroski, J, Wyatt, R, Nabel, G.J, Kwong, P.D. | | Deposit date: | 2006-11-20 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural definition of a conserved neutralization epitope on HIV-1 gp120.
Nature, 445, 2007
|
|
3SV5
 
 | | Engineered medium-affinity halide-binding protein derived from YFP: iodide complex | | Descriptor: | 1,2-ETHANEDIOL, FORMIC ACID, Green fluorescent protein, ... | | Authors: | Wang, W, Grimley, J.S, Beese, L.S, Hellinga, H.W. | | Deposit date: | 2011-07-12 | | Release date: | 2012-07-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Visualization of Synaptic Inhibition with an Optogenetic Sensor Developed by Cell-Free Protein Engineering Automation.
J.Neurosci., 33, 2013
|
|
3GWU
 
 | | Leucine transporter LeuT in complex with sertraline | | Descriptor: | (1S,4S)-4-(3,4-dichlorophenyl)-N-methyl-1,2,3,4-tetrahydronaphthalen-1-amine, LEUCINE, SODIUM ION, ... | | Authors: | Zhou, Z, Zhen, J, Karpowich, N.K, Law, C.J, Reith, M.E.A, Wang, D.N. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Antidepressant specificity of serotonin transporter suggested by three LeuT-SSRI structures.
Nat.Struct.Mol.Biol., 16, 2009
|
|
3GXF
 
 | |
3SUF
 
 | | Crystal structure of NS3/4A protease variant D168A in complex with MK-5172 | | Descriptor: | (1aR,5S,8S,10R,22aR)-5-tert-butyl-N-{(1R,2S)-1-[(cyclopropylsulfonyl)carbamoyl]-2-ethenylcyclopropyl}-14-methoxy-3,6-di oxo-1,1a,3,4,5,6,9,10,18,19,20,21,22,22a-tetradecahydro-8H-7,10-methanocyclopropa[18,19][1,10,3,6]dioxadiazacyclononadec ino[11,12-b]quinoxaline-8-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-11 | | Release date: | 2012-09-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SX0
 
 | | Crystal structure of Dot1l in complex with a brominated SAH analog | | Descriptor: | (2S)-2-amino-4-({[(2S,3S,4R,5R)-5-(4-amino-5-bromo-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}sulfanyl)butanoic acid (non-preferred name), Histone-lysine N-methyltransferase, H3 lysine-79 specific, ... | | Authors: | Yu, W, Tempel, W, Smil, D, Schapira, M, Li, Y, Vedadi, M, Nguyen, K.T, Wernimont, A.K, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-07-14 | | Release date: | 2011-07-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Bromo-deaza-SAH: a potent and selective DOT1L inhibitor.
Bioorg. Med. Chem., 21, 2013
|
|
3SV7
 
 | | Crystal structure of NS3/4A protease variant R155K in complex with Telaprevir | | Descriptor: | (1S,3aR,6aS)-2-[(2S)-2-({(2S)-2-cyclohexyl-2-[(pyrazin-2-ylcarbonyl)amino]acetyl}amino)-3,3-dimethylbutanoyl]-N-[(2R,3S)-1-(cyclopropylamino)-2-hydroxy-1-oxohexan-3-yl]octahydrocyclopenta[c]pyrrole-1-carboxamide, NS3 protease, NS4A protein, ... | | Authors: | Schiffer, C.A, Romano, K.P. | | Deposit date: | 2011-07-12 | | Release date: | 2012-09-05 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The Molecular Basis of Drug Resistance against Hepatitis C Virus NS3/4A Protease Inhibitors.
Plos Pathog., 8, 2012
|
|
3SXA
 
 | | Crystal structure of ABBB+UDP+Gal with Glycerol as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Histo-blood group ABO system transferase, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2O2U
 
 | | Crystal structure of human JNK3 complexed with N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Descriptor: | Mitogen-activated protein kinase 10, N-(3-cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)-2-fluorobenzamide | | Authors: | Somers, D, Rowland, P. | | Deposit date: | 2006-11-30 | | Release date: | 2007-02-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | N-(3-Cyano-4,5,6,7-tetrahydro-1-benzothien-2-yl)amides as potent, selective, inhibitors of JNK2 and JNK3.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
3SXG
 
 | | Crystal structure of AAAA+UDP+Gal with MPD as the cryoprotectant | | Descriptor: | DI(HYDROXYETHYL)ETHER, Histo-blood group ABO system transferase, MANGANESE (II) ION, ... | | Authors: | Johal, A.R, Evans, S.V. | | Deposit date: | 2011-07-14 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Sequence-dependent effects of cryoprotectants on the active sites of the human ABO(H) blood group A and B glycosyltransferases.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2O5E
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glucose pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', CHLORIDE ION, DNA topoisomerase 3, ... | | Authors: | Changela, A, DiGate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
2O4Q
 
 | | Structure of Phosphotriesterase mutant G60A | | Descriptor: | CACODYLATE ION, Parathion hydrolase, ZINC ION | | Authors: | Kim, J, Ramagopal, U.A, Tsai, P.C, Raushel, F.M, Almo, S.C. | | Deposit date: | 2006-12-04 | | Release date: | 2007-12-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of diethyl phosphate bound to the binuclear metal center of phosphotriesterase.
Biochemistry, 47, 2008
|
|
2O54
 
 | | Structure of E. coli topoisomerase III in complex with an 8-base single stranded oligonucleotide. Frozen in glycerol at pH 7.0 | | Descriptor: | 5'-D(*CP*GP*CP*AP*AP*CP*TP*T)-3', ACETIC ACID, CHLORIDE ION, ... | | Authors: | Changela, A, Digate, R.J, Mondragon, A. | | Deposit date: | 2006-12-05 | | Release date: | 2007-04-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Studies of E. coli Topoisomerase III-DNA Complexes Reveal a Novel Type IA Topoisomerase-DNA Conformational Intermediate.
J.Mol.Biol., 368, 2007
|
|
3H0S
 
 | |
3SZY
 
 | |
3H1V
 
 | | Human glucokinase in complex with a synthetic activator | | Descriptor: | 1-({5-[4-(methylsulfonyl)phenoxy]-2-pyridin-2-yl-1H-benzimidazol-6-yl}methyl)pyrrolidine-2,5-dione, Glucokinase, SODIUM ION, ... | | Authors: | Kamata, K, Takahashi, K. | | Deposit date: | 2009-04-14 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The design and optimization of a series of 2-(pyridin-2-yl)-1H-benzimidazole compounds as allosteric glucokinase activators.
Bioorg.Med.Chem., 17, 2009
|
|
3SYA
 
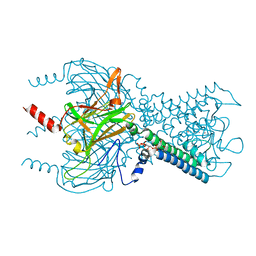 | | Crystal structure of the G protein-gated inward rectifier K+ channel GIRK2 (Kir3.2) in complex with sodium and PIP2 | | Descriptor: | G protein-activated inward rectifier potassium channel 2, POTASSIUM ION, SODIUM ION, ... | | Authors: | Whorton, M.R, MacKinnon, R. | | Deposit date: | 2011-07-16 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Crystal Structure of the Mammalian GIRK2 K(+) Channel and Gating Regulation by G Proteins, PIP(2), and Sodium.
Cell(Cambridge,Mass.), 147, 2011
|
|
3GXT
 
 | |
