7KAI
 
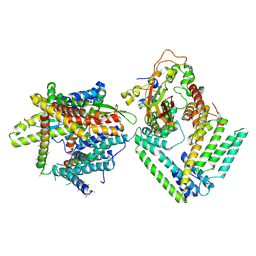 | | Cryo-EM structure of the Sec complex from S. cerevisiae, wild-type, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAU
 
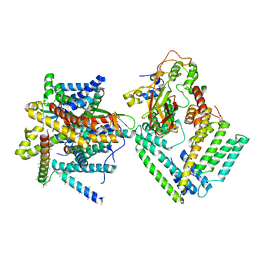 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore ring and Sec63 FN3 double mutant, class with Sec62 | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KAP
 
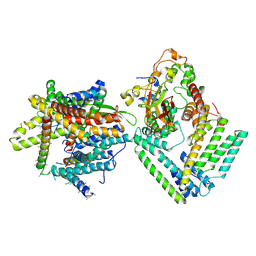 | | Cryo-EM structure of the Sec complex from S. cerevisiae, Sec61 pore mutant, class with Sec62, conformation 1 (C1) | | Descriptor: | Protein translocation protein SEC63, Protein transport protein SBH1, Protein transport protein SEC61, ... | | Authors: | Itskanov, S, Park, E. | | Deposit date: | 2020-10-01 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Stepwise gating of the Sec61 protein-conducting channel by Sec63 and Sec62.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7KB5
 
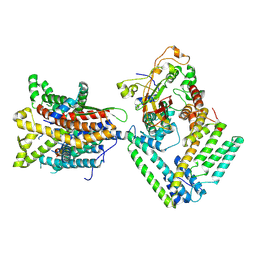 | |
7BV8
 
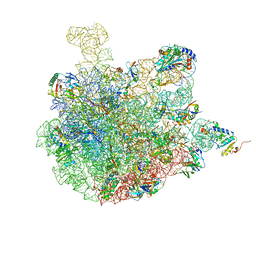 | | Mature 50S ribosomal subunit from RrmJ knock out E.coli strain | | Descriptor: | 23S rRNA, 50S ribosomal protein L10, 50S ribosomal protein L11, ... | | Authors: | Wang, W, Li, W.Q, Ge, X.L, Yan, K.G, Mandava, C.S, Sanyal, S, Gao, N. | | Deposit date: | 2020-04-09 | | Release date: | 2020-07-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.14 Å) | | Cite: | Loss of a single methylation in 23S rRNA delays 50S assembly at multiple late stages and impairs translation initiation and elongation.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5IQR
 
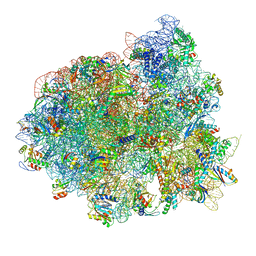 | | Structure of RelA bound to the 70S ribosome | | Descriptor: | 30S ribosomal protein S10, 30S ribosomal protein S11, 30S ribosomal protein S12, ... | | Authors: | Brown, A, Fernandez, I.S, Gordiyenko, Y, Ramakrishnan, V. | | Deposit date: | 2016-03-11 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Ribosome-dependent activation of stringent control.
Nature, 534, 2016
|
|
7BHP
 
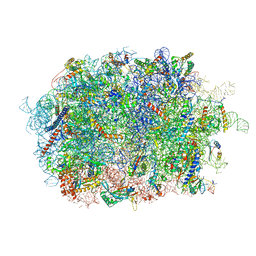 | | Cryo-EM structure of the human Ebp1 - 80S ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Desogus, J, Bhaskar, V, Chao, J.A. | | Deposit date: | 2021-01-11 | | Release date: | 2021-02-03 | | Last modified: | 2021-03-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Dynamic association of human Ebp1 with the ribosome.
Rna, 27, 2021
|
|
7BT6
 
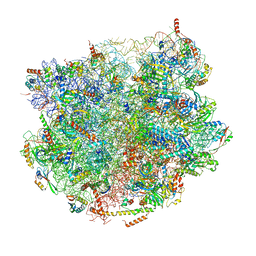 | |
7BTB
 
 | |
5GAH
 
 | | RNC in complex with SRP with detached NG domain | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
7K54
 
 | | Mid-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure II-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7AQR
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (peripheral arm) | | Descriptor: | Acyl carrier protein 2, mitochondrial, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-22 | | Release date: | 2021-12-08 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7K52
 
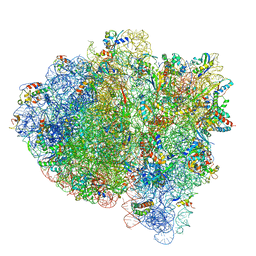 | | Near post-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure III) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
5GAG
 
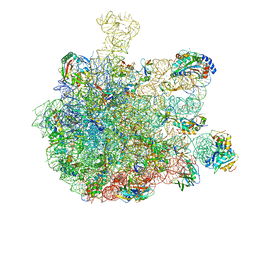 | | RNC in complex with SRP-SR in the closed state | | Descriptor: | 1A9L SS, 23S rRNA, 50S ribosomal protein L10, ... | | Authors: | Jomaa, A, Boehringer, D, Leibundgut, M, Ban, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structures of the E. coli translating ribosome with SRP and its receptor and with the translocon.
Nat Commun, 7, 2016
|
|
7ARC
 
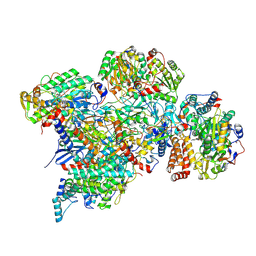 | | Cryo-EM structure of Polytomella Complex-I (peripheral arm) | | Descriptor: | 13 kDa, 18 kDa, 24 kDa, ... | | Authors: | Klusch, N, Kuehlbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7K53
 
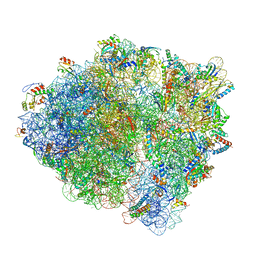 | | Pre-translocation +1-frameshifting(CCC-A) complex (Structure I-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K55
 
 | | Near post-translocated +1-frameshifting(CCC-A) complex with EF-G and GDPCP (Structure III-FS) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K51
 
 | | Mid-translocated non-frameshifting(CCA-A) complex with EF-G and GDPCP (Structure II) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7K50
 
 | | Pre-translocation non-frameshifting(CCA-A) complex (Structure I) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Demo, G, Loveland, A.B, Svidritskiy, E, Gamper, H.B, Hou, Y.M, Korostelev, A.A. | | Deposit date: | 2020-09-16 | | Release date: | 2021-07-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for +1 ribosomal frameshifting during EF-G-catalyzed translocation.
Nat Commun, 12, 2021
|
|
7AR8
 
 | | Cryo-EM structure of Arabidopsis thaliana complex-I (closed conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
7ARB
 
 | | Cryo-EM structure of Arabidopsis thaliana Complex-I (complete composition) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W, Yildiz, O. | | Deposit date: | 2020-10-23 | | Release date: | 2021-12-15 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
2WWA
 
 | | Cryo-EM structure of idle yeast Ssh1 complex bound to the yeast 80S ribosome | | Descriptor: | 25S RRNA, 60S RIBOSOMAL PROTEIN L17-A, 60S RIBOSOMAL PROTEIN L19, ... | | Authors: | Becker, T, Mandon, E, Bhushan, S, Jarasch, A, Armache, J.P, Funes, S, Jossinet, F, Gumbart, J, Mielke, T, Berninghausen, O, Schulten, K, Westhof, E, Gilmore, R, Beckmann, R. | | Deposit date: | 2009-10-22 | | Release date: | 2009-12-08 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Structure of Monomeric Yeast and Mammalian Sec61 Complexes Interacting with the Translating Ribosome.
Science, 326, 2009
|
|
5IT8
 
 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-03-16 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.12 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7AR7
 
 | | Cryo-EM structure of Arabidopsis thaliana complex-I (open conformation) | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, (7S)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, ... | | Authors: | Klusch, N, Kuelbrandt, W. | | Deposit date: | 2020-10-23 | | Release date: | 2022-03-02 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | A ferredoxin bridge connects the two arms of plant mitochondrial complex I.
Plant Cell, 33, 2021
|
|
2X1X
 
 | | CRYSTAL STRUCTURE OF VEGF-C IN COMPLEX WITH DOMAINS 2 AND 3 OF VEGFR2 IN A TETRAGONAL CRYSTAL FORM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MERCURY (II) ION, ... | | Authors: | Leppanen, V.-M, Prota, A.E, Jeltsch, M, Anisimov, A, Kalkkinen, N, Strandin, T, Lankinen, H, Goldman, A, Ballmer-Hofer, K, Alitalo, K. | | Deposit date: | 2010-01-08 | | Release date: | 2010-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Determinants of Growth Factor Binding and Specificity by Vegf Receptor 2.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
