7ROH
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 11 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROG
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 10 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROC
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 7 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7ROD
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 8 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
6M8Y
 
 | | PSEUDOMONAS SERINE-CARBOXYL PROTEINASE (SEDOLISIN) COMPLEXED WITH THE INHIBITOR AIPF | | Descriptor: | AIPF PEPTIDE INHIBITOR, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Wlodawer, A, Li, M, Gustchina, A, Dauter, Z, Uchida, K, Oyama, H, Goldfarb, N.E, Dunn, B.M, Oda, K. | | Deposit date: | 2018-08-22 | | Release date: | 2018-10-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Inhibitor complexes of the Pseudomonas serine-carboxyl proteinase
Biochemistry, 40, 2001
|
|
5JSR
 
 | |
7ROE
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 9 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
6MA2
 
 | | Crystal structure of human O-GlcNAc transferase bound to a peptide from HCF-1 pro-repeat 2 (11-26) and inhibitor ent-1a | | Descriptor: | Host Cell Factor 1 peptide, N-[(2S)-2-(2-methoxyphenyl)-2-{[(2-oxo-1,2-dihydroquinolin-6-yl)sulfonyl]amino}acetyl]-N-[(thiophen-2-yl)methyl]glycine, UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 kDa subunit | | Authors: | Martin, S.E.S, Lazarus, M.B, Walker, S. | | Deposit date: | 2018-08-25 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Evolution of Low Nanomolar O-GlcNAc Transferase Inhibitors.
J. Am. Chem. Soc., 140, 2018
|
|
6UXR
 
 | | Crystal structure of BAK core domain BH3-groove-dimer in complex with LysoPC | | Descriptor: | Bcl-2 homologous antagonist/killer, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL, ... | | Authors: | Cowan, A.D, Colman, P.M, Czabotar, P.E. | | Deposit date: | 2019-11-07 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | BAK core dimers bind lipids and can be bridged by them.
Nat.Struct.Mol.Biol., 27, 2020
|
|
5MRG
 
 | | Solution structure of TDP-43 (residues 1-102) | | Descriptor: | TAR DNA-binding protein 43 | | Authors: | Mompean, M, Romano, V, Pantoja-Uceda, D, Stuani, C, Baralle, F.E, Laurents, D.V. | | Deposit date: | 2016-12-22 | | Release date: | 2017-06-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Point mutations in the N-terminal domain of transactive response DNA-binding protein 43 kDa (TDP-43) compromise its stability, dimerization, and functions.
J. Biol. Chem., 292, 2017
|
|
7RO5
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 4 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
8B2T
 
 | | SARS-CoV-2 Main Protease (Mpro) in complex with nirmatrelvir alkyne | | Descriptor: | 3C-like proteinase nsp5, Nirmatrelvir (reacted form) | | Authors: | Owen, C.D, Crawshaw, A.D, Warren, A.J, Trincao, J, Zhao, Y, Brewitz, L, Malla, T.R, Salah, E, Petra, L, Strain-Damerell, C, Schofield, C.J, Walsh, M.A. | | Deposit date: | 2022-09-14 | | Release date: | 2023-02-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.893 Å) | | Cite: | Alkyne Derivatives of SARS-CoV-2 Main Protease Inhibitors Including Nirmatrelvir Inhibit by Reacting Covalently with the Nucleophilic Cysteine.
J.Med.Chem., 66, 2023
|
|
7ROB
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 6 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
7RO6
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 5 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
6BWH
 
 | |
7RO3
 
 | | Cryo-EM reconstruction of Sulfolobus monocaudavirus SMV1, symmetry 2 | | Descriptor: | major capsid protein | | Authors: | Wang, F, Cvirkaite-Krupovic, V, Krupovic, M, Egelman, E.H. | | Deposit date: | 2021-07-30 | | Release date: | 2022-03-30 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Spindle-shaped archaeal viruses evolved from rod-shaped ancestors to package a larger genome.
Cell, 185, 2022
|
|
8JU3
 
 | | Mu phage tail fiber | | Descriptor: | Tail fiber protein S,Tail fiber protein S' | | Authors: | Yamashita, E, Takeda, S. | | Deposit date: | 2023-06-24 | | Release date: | 2024-02-28 | | Last modified: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Determination of the three-dimensional structure of bacteriophage Mu(-) tail fiber and its characterization.
Virology, 593, 2024
|
|
8CPB
 
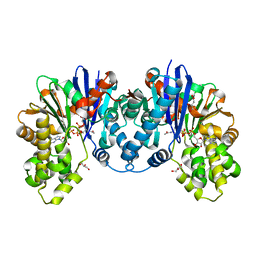 | | 1,6-anhydro-n-actetylmuramic acid kinase (AnmK) in complex with AMPPNP, and AnhMurNAc at 1.7 Angstroms resolution. | | Descriptor: | 2-(2-ACETYLAMINO-4-HYDROXY-6,8-DIOXA-BICYCLO[3.2.1]OCT-3-YLOXY)-PROPIONIC ACID, Anhydro-N-acetylmuramic acid kinase, GLYCEROL, ... | | Authors: | Jimenez-Faraco, E, Hermoso, J.A. | | Deposit date: | 2023-03-02 | | Release date: | 2023-09-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Catalytic process of anhydro-N-acetylmuramic acid kinase from Pseudomonas aeruginosa.
J.Biol.Chem., 299, 2023
|
|
6MBE
 
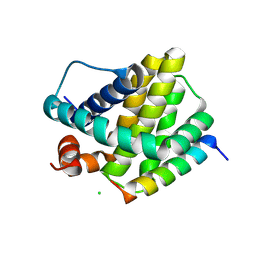 | | Human Mcl-1 in complex with the designed peptide dM7 | | Descriptor: | CHLORIDE ION, Induced myeloid leukemia cell differentiation protein Mcl-1, dM7 | | Authors: | Jenson, J.M, Keating, A.E. | | Deposit date: | 2018-08-29 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Tertiary Structural Motif Sequence Statistics Enable Facile Prediction and Design of Peptides that Bind Anti-apoptotic Bfl-1 and Mcl-1.
Structure, 27, 2019
|
|
6R6H
 
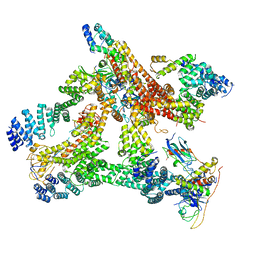 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
6IDL
 
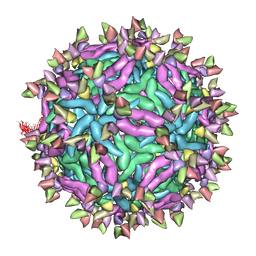 | | Cryo-EM structure of Immature Dengue virus serotype 3 in complex with human antibody 1H10 Fab at pH 5.0 (Class II particle) | | Descriptor: | Envelope protein, Fab 1H10 heavy chain (V-region), Fab 1H10 light chain (V-region), ... | | Authors: | Wirawan, M, Fibriansah, G, Ng, T.S, Zhang, Q, Kostyuchenko, V.A, Shi, J, Lok, S.M. | | Deposit date: | 2018-09-10 | | Release date: | 2018-12-12 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Mechanism of Enhanced Immature Dengue Virus Attachment to Endosomal Membrane Induced by prM Antibody.
Structure, 27, 2019
|
|
5N49
 
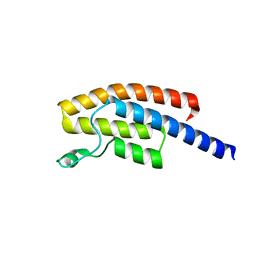 | | BRPF2 in complex with Compound 7 | | Descriptor: | 2-(1,3,6-trimethyl-2-oxidanylidene-benzimidazol-5-yl)benzo[de]isoquinoline-1,3-dione, Bromodomain-containing protein 1 | | Authors: | Bouche, L, Christ, C.D, Siegel, S, Fernandez-Montalvan, A.E, Holton, S.J, Fedorov, O, ter Laak, A, Sugawara, T, Stoeckigt, D, Tallant, C, Bennett, J, Monteiro, O, Saez, L.D, Siejka, P, Meier, J, Puetter, V, Weiske, J, Mueller, S, Huber, K.V.M, Hartung, I.V, Haendler, B. | | Deposit date: | 2017-02-10 | | Release date: | 2017-05-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Benzoisoquinolinediones as Potent and Selective Inhibitors of BRPF2 and TAF1/TAF1L Bromodomains.
J. Med. Chem., 60, 2017
|
|
5N2A
 
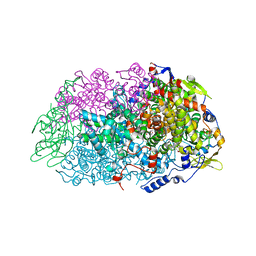 | | METHYL-COENZYME M REDUCTASE III FROM METHANOTORRIS FORMICICUS TRIGONAL FORM | | Descriptor: | 1-THIOETHANESULFONIC ACID, BROMIDE ION, Coenzyme B, ... | | Authors: | Wagner, T, Wegner, C.E, Ermler, U, Shima, S. | | Deposit date: | 2017-02-07 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Phylogenetic and Structural Comparisons of the Three Types of Methyl Coenzyme M Reductase from Methanococcales and Methanobacteriales.
J.Bacteriol., 199, 2017
|
|
8C54
 
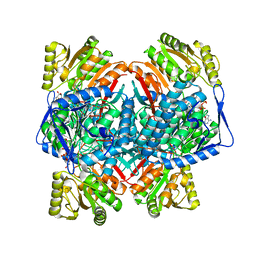 | | Cryo-EM structure of NADH bound SLA dehydrogenase RlGabD from Rhizobium leguminosarum bv. trifolii SRD1565 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Succinate semialdehyde dehydrogenase | | Authors: | Sharma, M, Meek, R.W, Armstrong, Z, Blaza, J.N, Alhifthi, A, Li, J, Goddard-Borger, E.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2023-01-06 | | Release date: | 2023-09-20 | | Last modified: | 2024-04-10 | | Method: | ELECTRON MICROSCOPY (2.52 Å) | | Cite: | Molecular basis of sulfolactate synthesis by sulfolactaldehyde dehydrogenase from Rhizobium leguminosarum.
Chem Sci, 14, 2023
|
|
5K3F
 
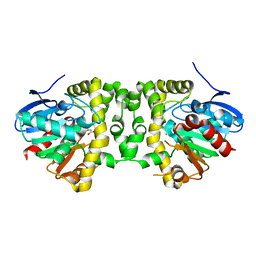 | | Crystal Structure of the Fluoroacetate Dehalogenase RPA1163 - His280Asn/Fluoroacetate - Cocrystallized - Single Protomer Reacted with Ligand | | Descriptor: | CHLORIDE ION, Fluoroacetate dehalogenase | | Authors: | Mehrabi, P, Kim, T.H, Prosser, S.R, Pai, E.F. | | Deposit date: | 2016-05-19 | | Release date: | 2017-02-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | The role of dimer asymmetry and protomer dynamics in enzyme catalysis.
Science, 355, 2017
|
|
